The incidence of autism spectrum disorders has increased sharply since it was first described 60 years ago.
Today, ASD affects more than 1% of all children in the U.S. and about half of them develop a life-long disability.
The overall cost to society is $100 billion per year, yet there is no treatment available to change to course of the disease.
What is known about the causes of the disease and how has your research added to this knowledge?
A rapid increase in incidence over only two generations means that environmental factors must play a role, yet no such factors have been identified. On the other hand, autism runs in families, so genetics must also play an important role.
Studies in families have identified hundreds of genes, yet it is unknown which of these genes are common risk factors and no connected pathways have been identified that could guide the development of treatments.
Most population-based studied have used statistical methods that focus on a single position (letter) among the 3 million positions with vacations (SNPs) along the chromosome.
If an isolated variation would increase the risk for autism, however, it would be selected against.
Hence, we have developed a novel statistical approach that looks for neighboring SNPs (words) where combinations of misspelled words convey risk, even if each of the letters, by itself, carries only a minor risk or may even be beneficial.
What are the benefits of this approach?
The approach we have developed avoids creating artefacts by making fewer assumptions than most traditional approaches, which may just count rare variations at different SNPs (apples and oranges). It also requires more memory than computers had until about 10 years ago and hundreds of computers equipped with massively parallel (GPU) processors.
Please can you outline the link between autism and epilepsy and how this links to Ras/calcium signaling?
The first application of this wide-locus GWAS in childhood absence epilepsy confirmed most of the known drug targets based on data of 185 children only. Most of the genes were clustered around ion channels and the "Ras" pathway involved in growth regulation. Traditional single-SNP GWAS, in contrast, had identified only a single locus with unknown function.
Since autism and epilepsies run in the same families, we then hypothesized that Ras/calcium signaling would also be involved in autism, which is broadly consistent with the known results from family studies.
Because the wide-locus approach requires fewer subjects to produce conclusive results, we analyzed the two "stages" of a publicly available data set separately and compared more severe vs less severe cases.
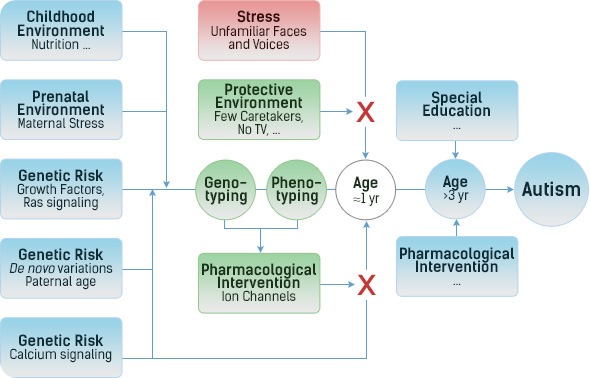
What did your analysis find?
Both analyses confirmed the involvement of the Ras/calcium pathway and both analyses showed one additional cluster, which contains several genes (PTPRs) involved in the down-regulation of growth factor receptors.
With autism being a neuro-developmental disease, this implied overgrowth of neurons as a critical component in the etiology of more severe forms of autism and, thus, the first two years of life as the window of opportunity for a pharmaceutical intervention.
A separate analysis, comparing all cases together with controls from a different study didn't show any differences in PTPRs between ASD and controls. That suggests that too much growth, by itself, does not cause autism, but merely larger brains and bodies.
Similarly, high levels of excitatory ions, by themselves, may cause epilepsies, but not autism. It is the combination of overgrowth and overexcitation (in response to environmental stress) that causes the more severe forms of autism.
Increases in environmental stress over the last 50 years, such as urbanization and TV, might have contributed to the increase in incidence in children with these genetic risk factors.
Giving small children drugs that down-regulate growth factors (Gleevac) would interfere with growth, in general. From epilepsies and rheumatoid arthritis, however, we know that down-regulating excitatory ion channels is well tolerated, in general. Hence, modulation of ion channels in children at the age of about 12 months, when the first symptoms of autism can be detected, may prevent progression to the more severe end of the spectrum.
Can this approach be applied to other conditions?
Of course, this novel computational biostatistics approach is not limited to neurodevelopmental diseases. Our results suggest that a relevant portion of genetic risk for common diseases is determined through coding variations — as was widely expected after deciphering the human genome ten years ago — and could be detected, but only with a statistical approach better adapted to the specifics of common diseases.
Reanalyzing the data already collected, including data from publicly available repositories (such as the NIH’s dbGaP), could finally lead to the insights sought for and the therapies urgently needed.
With the sample size requirements reduced from tens of thousands to a few hundred, subgroup analyses of "failed" phase III studies may further increase the number of treatment options by identifying the genetic predictors for patients most likely to benefit from a drug.
Where can readers find more information?
The results are published in the Nature Group journal Translational Psychiatry: http://www.nature.com/tp/journal/v4/n1/abs/tp2013124a.html
The computational biostatistics infrastructure is available via a Web site (https://www.rockefeller.edu/), where scientists can upload their data for analysis.
About Dr Knut M. Wittkowski
 Dr. Wittkowski received his MS in Statistics at the University of Carl-Friedrich Gauss, Göttingen, his PhD in Informatics at the Technical University of Stuttgart, and his ScD in Medical Biometry at the Eberhard-Carls-University, Tübingen.
Dr. Wittkowski received his MS in Statistics at the University of Carl-Friedrich Gauss, Göttingen, his PhD in Informatics at the Technical University of Stuttgart, and his ScD in Medical Biometry at the Eberhard-Carls-University, Tübingen.
An internationally renowned scientist with more than 30 years of work at the interface of non-parametric statistics and computer science, he is a leader in the emerging field of computational biostatistics and research design.
After joining The Rockefeller University, he worked on a class of non-parametric statistical methods whose application to multivariate data had been abandoned in the 1940s because of high memory demand.
His expansion of u-statistics to reflect structures among variables has been successfully applied to a wide range of applications, including sports, consumer preferences, and images, as well as complex phenotypes in Fanconi anemia, psoriasis, shock, epilepsies, autism, and many other diseases.
He is the author of 63 publications and 7 patents, including one on improving personalized medicine through the use of hierarchically structured u-statistics to integrate information from complex phenotypes, genetically structured data, and gene-expression profiles.