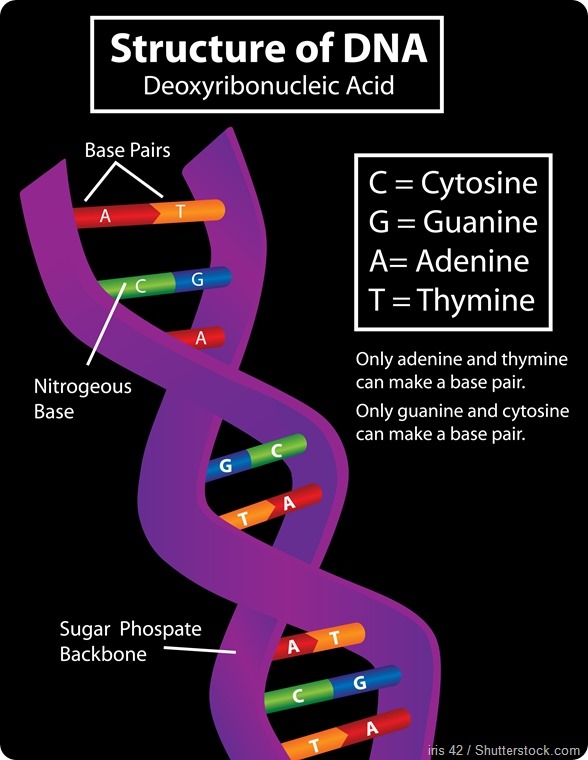
The natural DNA bases that form the letters of DNA are usually referred to as G, C, A, and T. Those are only the first letters of the chemical names. They’re often called nucleotides by their scientific name and all of them have in common a phosphate part, a sugar part and a nucleobase part.
In all of G, C, A, and T the phosphate and the sugar part are the same; what differentiates them is their nucleobase part. Those nucleobase parts are what pair with each other: the nucleobase of G interacts with the nucleobase of C and that’s why G and C pair together; the nucleobase of A forms hydrogen bonds with the nucleobase of T and that’s why A and T pair together.
That pairing is what makes DNA. A always pairs with T and G always pairs with C. They pair with each other through complementary hydrogen bond formation.
A hydrogen bond is an interaction where two atoms share one hydrogen atom. They’re each holding onto the same hydrogen atom, and that provides a force to interact with each other through.
What is thought to be the reason why organisms only use two pairs of DNA bases and what sparked your interest in creating an organism that contains an additional ‘unnatural’ pair of bases?
That’s something my lab is really interested in and has been a fundamental question in science and evolution: why four letters and two base pairs? Why 20 amino acids?
Why life is the way it is today is a question people have been asking and speculating about for years. The most fundamental form of that question is the letters of DNA—G, C, A, and T—and those two base pairs, because that’s the system that stores information.
All the diversity that life can draw upon is encoded in those letters, so this is probably as fundamental an area in which to ask that question as any.
I think that, for a long time, there was a suspicion that the natural letters and the two natural base pairs were the only possible solution to storing information. For example, if life would have evolved elsewhere it would have had to use the same letters; however, our recent work shows that’s really not true and there certainly are other solutions, maybe even lots of other solutions.
There is really nothing magic about G, C, A, and T: you just need suitable forces that underlie their stable pairing with each other, and their stable recognition of each other, during replication by DNA polymerase.
What this shows is that, at least for DNA, there were other solutions possible and G, C, A, and T are not uniquely special in that regard.
In a lot of ways, what we’ve done is make a small change to DNA: the natural nucleotides all have the same phosphate and sugar parts and are differentiated by their nucleobase; we have the same phosphate, the same sugar and, just like G, C, A, and T, all we changed was the nucleobase part so that our new nucleotides, X and Y, pair with each other.
Their nucleobases recognize each other and they don’t recognize any of the natural nucleobases, so the nucleobases are the only thing that differentiates our letters from G, C, A, and T.
Scientists are conservative by nature, and they should be, but if DNA was able to make a small change and still store information, maybe a big change is possible; maybe bigger changes to DNA could have been made and you could have still stored information that way.
In fact, if you really wanted to be speculative, if small change is possible then maybe really big changes are possible and you would not have needed to use DNA at all.
Our new letters, X and Y, are similar to G, C, A, and T in a lot of ways, but I want to emphasize that, from another perspective, they’re actually quite different because G, C, A, and T pair with each other based on the formation of complementary hydrogen bonds. Our base pairs don’t interact by hydrogen bonds; they interact by a completely different force.
They interact by something that scientists call the hydrophobic effect, so they’re oily. Everyone is familiar with the fact that oil and water don’t mix. The natural bases are able to form hydrogen bonds and that’s very water-like.
We designed our pairs to be very fatty, or oily, so that they wouldn’t pair with water; they wouldn’t interact with water and they also wouldn’t interact with the water-like hydrogen bonds of the natural nucleobases.
From that perspective our unnatural base pairs really are quite different because they’re not just a subtle variation on the same theme: they draw on a completely different force for recognition and for formation of the X-Y base pair.
From one perspective they’re similar, they’re like natural DNA because they have the same phosphorous and the same sugar, but their nucleobases are hydrophobic and the mechanism of interacting with each other is really quite different.
How do the two forces, the hydrogen bonds and hydrophobic interactions, compare in terms of strength?
That’s a complicated question, which people have been asking for decades. The hydrophobic force certainly contributes to protein structure, so people have been arguing and thinking about what the varying strength of a hydrogen bond is, relative to hydrophobic interactions.
Hydrophobic interactions are a little bit more difficult to describe and think about because they’re not a single force. We tend to use the term hydrophobic effect a bit loosely.
Technically, what the hydrophobic effect refers to is the fact that oil wants to get out of water. That’s a big part of what drives the stability of our unnatural base pair, because its only other option is to be in water.
There are other forces involved, so the purists who think about this would object to our simply calling them the hydrophobic effect. When you pack molecules on each other, aromatic molecules, like our nucleobases, interact with each other in a stabilizing way; they pack on each other very nicely, in a way that stabilizes their interaction. That’s in addition to the hydrophobic effect so, when we write papers and describe our unnatural base pairs, we usually describe them as pairing based on hydrophobic and packing forces.
When you pack these aromatic molecules on each other it tends to be stabilizing, so you have to add that into it. As far as which is stronger that’s a tough question because the hydrophobic interactions we’re drawing on are not one single thing; they’re a composite of things and they certainly would be much weaker in water.
Part of our design criteria during the development of the unnatural base pair was to put the unnatural base pair into duplex DNA and measure its stability. We always had things that were close to being as stable as the natural base pairs.
A lot of times people will call the hydrophobic effect a little bit weaker than hydrogen bonding but, when you add up all the contributions with our unnatural base pair, they somehow add up to X-Y being as good as a natural base pair because we don’t destabilize the duplex.
Please can you outline exactly what this work has involved?
My lab first started about 15 years ago, around 1999, with the idea of developing an unnatural base pair. From the very beginning, we thought that selectivity of pairing would be a critical factor: when you have X in DNA and you’re copying it, to replicate it to pass onto the next generation, you have to only incorporate Y; you can’t incorporate G, C, A, or T. That would be a mutation.
Likewise, when you have Y in the template DNA that you’re copying you have to only incorporate X; again, you can’t incorporate G, C, A, or T.
We thought that selectivity, to favor X pairing with Y and not pairing with G, C, A, or T, would be one of the biggest challenges. That’s why, from the beginning, we thought we would use hydrophobic and packing forces. No matter what your design is, oil and water don’t want to mix so it would buy us some orthogonality, some specificity against mispairing, with the natural nucleobases if we made them hydrophobic.
We designed several scaffolds, where there were just these nucleobases of very simple rings, and we started decorating those rings with hydrophobic groups. It very empirically followed to progress the project by making analogues, characterizing them and then slowly optimizing them.
The way we would optimize them is we would characterize them in a test tube: we would put our unnatural, X-like nucleotide in a template and then we would determine the ability of the DNA polymerase to replicate it in the test tube. This is a much simpler environment than a cell.
We would also characterize its stability in duplex DNA. We’d make a new analogue, characterize it and then, if we made some modification that made those properties better, we would note that and we would use that as a design criteria in the future. If we made a modification that made it worse, made its recognition by DNA polymerase worse, for example, then we would tend not to use that type of modification.
Over 12-13 years we kept designing more and more analogues, finding which things made them better, combining those things in new ways and trying to continue to make them better. In the end, we probably approached 300 different nucleotide analogues and all of them had these hydrophobic nucleobases. All of them were characterized by looking at their interactions with each other, trying to find two that selectively paired with each other and that didn’t pair with G, C, A, or T, both in the duplex and also during replication by DNA polymerase.
DNA polymerases are the proteins in the cell that are responsible for replicating DNA. We also looked at RNA polymerases because any base pair that we get eventually has to be officially transcribed into RNA.
The way information flow goes in a cell is that you have this sequence of letters, your nucleotides of DNA, and that has to be copied into RNA. That RNA sequence determines protein sequence. One of the first steps of retrieving information within a cell is copying the DNA into RNA, so we wanted to make sure that our unnatural base pair was both efficiently replicated by DNA polymerases and efficiently transcribed into RNA by RNA polymerases.
It was this long, 12-13 year optimization, where we just kept making new analogues and characterizing them, that ultimately resulted in the identification of X-Y, which is a pair that forms really well with each other and not with G, C, A, or T.
What would happen if you included other hydrophobic bases?
That would probably be a very different challenge than what we’ve accomplished now. The very fact that it’s hydrophobic bought us a lot of selectivity against pairing with G, C, A, and T: if we made more hydrophobic analogues, pursuing the same strategy that we used to develop X and Y, one of the further constraints would be that these new letters would have to not pair with X and Y.
While no oil wants to mix with water, oil will mix with different oil quite well. It would be a rather different challenge. It’s certainly an interesting one, and one that I don’t think would be impossible, but it’s not something that my lab is working on.
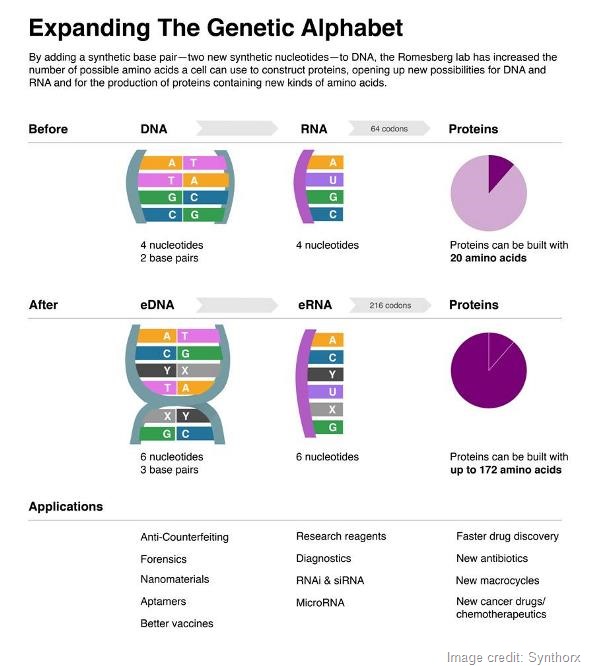
In the end, if all you want is to simply store additional information in a cell and retrieve it, one unnatural base pair is sufficient because you can put it into different combinations of sequences. It will encode about ten times more information than we could ever even imagine using.
The increase in information is not linear, it’s nonlinear. The way information storage works is that the sequence of DNA is converted into a specific sequence of RNA and RNA is then read during protein synthesis in groups of three nucleotides, called codons.
GGA is a natural codon that would code for a natural amino acid. ATG, for example, codes for the methionine amino acid.
Because the words that we convert into proteins are comprised of three letters, you can arrange those letters in all sorts of combinations. The number of codons that are available, meaning in principle the number of unique amino acids that are available, form a four-letter, two-base-pair code is 64.
The first, second and third positions could be any of the four letters, so four letters allows for 64 different codons.
That’s not quite how it works because some of those codons are reserved for other things like signals that tell the protein synthesis to stop, for example. Also, the third position isn’t always specific, in a lot of cases, so you can have redundancy there.
In the end, those 64 possible codons encode for 20 amino acids.
If you have six letters, three base pairs and those same three positions, you instead have 216 possible codons. That’s more amino acids than we would ever be able to use.
While the idea of making yet another base pair is really interesting from a conceptual perspective, and these conceptual questions are a big part of what has driven this project in my lab, we’re also driven by practical goals like expressing proteins.
At the first level, we’ve already answered the question of whether an additional base pair is possible; what we want to do now is retrieve that information. We’ve shown that we can get cells to store more information in their DNA and now we want to take that even further by developing cells that retrieve that information in the way of RNA, and in the way synthesizing proteins that are comprised of not only the 20 natural amino acids, but say 21-25.
How did you stop natural DNA-repair mechanisms from removing the unnatural nucleosides and what other hurdles did you have to overcome in order to develop a new functional pair of DNA bases?
It’s something we worried a lot about. Every time I would write an NIH grant—the NIH is where you have to write grants to get funding to do research—I would talk about this project and I would always get that question from the reviewers. They thought it was interesting but that there’s a very good chance that, even if you could replicate the DNA, the DNA repair machines would interfere.
There’s also redundancy and multiple different repair pathways: nucleotide excision repair, base excision repair, methyl-directed mismatch repair, and there are glycosylases that recognize damaged DNA and cut it out. There are all sorts of repair mechanisms and they do things like recognize abnormal DNA, so it was certainly a concern of ours and a concern of every reviewer who reviewed our grants at the NIH.
My standard response was always that these repair pathways are nonessential: you can delete the genes, you can disable them, and so we would make cells that simply worked without those pathways.
The challenge is that that’s not a great solution because, if you had to delete all of them, the cell would be quite sick. So while that was my standard response, it was actually not a very attractive response.
This effort to go into a cell started about a year to a year and a half ago. We immediately envisioned three major challenges: one was getting the triphosphates, the activated forms of X and Y required for DNA synthesis, into a cell because the cell is not going to make them; you have to provide them.
The second challenge was whether, once you get them into a cell, natural DNA polymerase, the natural replication machinery of a cell, would be able to recognize and use them for DNA synthesis. That involves whether the X and Y substrate would even get to the right place within the cell and be recognized efficiently. One could imagine all sorts of problems coming up: they could get degraded or they could get sequestered away in the wrong part of the cell.
The third challenge was exactly what you just mentioned: even if you could get them into DNA, would they be removed by one or more of the different DNA repair pathways?
The first challenge turned out to be a huge challenge that we had to work on for about a year, but we finally got it to work. We thought the next two were going to be the big challenges, they would be even harder, and they turned out to utterly disappear as a challenge. They simply didn’t matter.
In retrospect, that’s the remarkable part of how this project went. Once we got the activated forms, the triphosphates, of X and Y into the cell, the standard replication machinery of the cell loved it; they replicated the DNA with really high efficiency and fidelity. I ascribe that to 12 years of optimization in the test tube.
The third challenge, the DNA repair pathways, they simply didn’t recognize them or cut them out. In our earliest experiments, and we’ve done this many times now, we propagated the bacteria over 15-25 hours, which corresponds to 20-30 generations, and the unnatural base pair was retained at a really high level. That tells you that, during the normal growth and division of a bacterial cell, these repair pathways were not removing them.
That might not be the whole story, because bacteria have largely two types of growth states: one is called exponential growth, where they’re growing very fast and doubling as fast as they can because you’ve given them lots of nutrients and they don’t care about anything else but growing; the other is called stationary phase and that’s where the cells slow down and simply stop growing because they’ve exhausted all the nutrients and are just waiting.
There’s some data in the literature that says a lot of these repair pathways might be preferentially turned on during the stationary phase. To explore that in this paper, we provided one batch of these required X and Y triphosphate substrates to the medium.
The triphosphates have a lifetime: they decompose and are about half gone by nine hours, and they’re 75% gone by about eighteen hours. That’s called a half-life, the time it takes for half to disappear, so they have a half-life of about nine hours.
By two or three days the triphosphates are basically completely gone, so we grew them up in the stationary phase and never provided more triphosphates; we let them just sit there in the absence of our unnatural X and Y triphosphates. There’s no way they can replicate, but if they would be cut out during the stationary phase you would lose them very quickly and you would see them suddenly go away.
What we found was that the loss of the unnatural base pair paralleled growth. You still get a little bit of growth during the stationary phase and the loss of the unnatural base pair exactly mirrored that growth and, on top of that, the way that you lost it suggested it was not lost due to repair.
A lot of times, if you lost it by any of these repair pathways, there are certain signatures and what we know from all that test tube work that we did was, when you lose it by forced mispairing in a polymerase, you replace it with an A-T. That’s the preference it has.
If you really force a DNA polymerase, you don’t give it the unnatural triphosphates, they’ll just sit there and eventually insert an A-T pair.
Over 3-5 days the loss of the unnatural base pair exactly paralleled the growth curve and it was replaced with an A-T. Every indication is that you do eventually lose it, which had to happen because we weren’t providing them any longer, but the way you lose them is by polymerases eventually making errors.
In addition, even after six days of sitting there with no triphosphates, 20% of the unnatural base pair was still there. What this shows is that the DNA repair pathways are not efficiently cutting them out, even during the stationary phase.
From a technical perspective, they are different from natural DNA and I think it’s entirely possible that some or all of these repair pathways do recognize it but can’t do anything about it, because it’s an unnatural base pair. They don’t have anything to grab onto. Maybe they’re not recognized, and this is something we’re interested in looking at in the future, but right now all that we know is that they’re not efficiently cut out by the repair pathways.
What happens when the new bases are not provided?
That’s actually an important issue because a lot of times I get questions like: isn’t this kind of scary? Shouldn’t people be scared of this because giving bacteria the ability to store more information might make them more fit, more dangerous?
This is an important question because what synthetic biology works on is the manipulation of life in a synthetic way, which means a lot of work that’s been done is just reorganizing the natural components of a pathway. When you do that there’s always the fear that something unexpected could happen because the cell always maintains all of the raw material it needs to reorganize, undo or modify what you’ve done in some unanticipated way.
A difference with our work is that X and Y are completely unnatural; they’re not found in nature or in a cell. They’re not anywhere. A lot of times people think it’s arrogant to say that nature couldn’t make X and Y because of scenarios like those in Jurassic Park; nature finds a way.
X and Y, the unnatural nucleotides, are so different from anything nature makes; they’re just nowhere near anything that nature could make and evolution doesn’t work by creating huge, complex, multistep pathways out of nothing.
For technical reasons I don’t want to go into this too much, but one of our unnatural base pairs, X, is a little bit more different from a natural nucleotide than Y. Most of the time, in these nucleotides, the nucleobase portion is attached to the sugar portion by a carbon-nitrogen bond. The reason it does that is because that’s a relatively easy bond to make in a cell.
One of our analogues is attached by a carbon-carbon bond. That is extraordinarily difficult to make because the intermediates are very unstable. In fact, nature doesn’t make anything like that, so the notion that nature could suddenly find a way to make this is maybe good science fiction, maybe a good story for Hollywood, but it’s far from what anyone would expect in reality.
If someone tried to grow this outside of the controlled environment of the lab, where you are providing X and Y to the medium, they would simply lose the unnatural base pairs. There’s really no way that they could suddenly find a way to create them, use them and have a cell that was viable so, if they did try to grow in the absence of the triphosphates, they revert back to natural; X-Y will eventually revert to an A-T.
The very synthetic component that makes them different, that gives them the increased information, will simply vanish.
That’s an advantage and a disadvantage: it’s an advantage for the reason it provides a fail-safe; it’s a disadvantage because it means we have to provide the bacteria with the unnatural letters.
We don’t really consider that a big deal. We consider it sort of as an additional nutrient, an additional vitamin, which we have to provide the cells. When you grow cells you always have to provide them with a lot of things: sugars, phosphates, nitrogen, a carbon source, metals, and all sorts of things. Our semi-synthetic cells have the additional nutrient requirements of adding the triphosphates of X and Y.
What impact do you hope this research will have?
It gets back to what our original motivations were. There are two motivations and I think that they’ll have their own impacts. The first is this conceptual notion that, if you look around nature anywhere in the world, in all the diversity that you see, from the lowest, simplest single-celled organisms all the way up to the most complex organisms like you and me, all of the information is encoded in a four-letter alphabet. That’s all the information that nature has to draw upon. That’s all that evolution has to draw upon.
Evolutionary biologists have a way of looking back in time and it appears that, all the way back to the last common ancestor of all life on Earth, it had a four letter alphabet. Going to six letters and showing that it’s possible has conceptual implications for our understanding of information storage in a cell and, therefore, our understanding of what life can be, because the information stored in a genome defines what life can be. I hope it impacts people thinking in that way, and I hope that some evolutionary biologists and people who think about evolution will consider it.
For people who think about the origins of life or why life evolved the way it evolved, this is a rare piece of experimental data that actually addresses that question.
The second impact, and this is maybe what I’m personally most excited about, is that my lab is going to focus on using the increased information and retrieving it to synthesize ultimately novel proteins.
Proteins have become a really big deal as therapeutics. It’s remarkable that, traditionally, therapeutics have been small molecules but lately, in the past 10 or so years, there’s been a revolution in drug discovery where people are using proteins.
In fact, last year, more novel candidate drugs that were announced were proteins than small molecules. That just shows this amazing switch towards proteins.
The disadvantage proteins have is that they can only be comprised of the 20 natural amino acids. What better drug behavior, what more amazing properties, could you get if they could be composed of 25?
You could maybe add diversity, chemical structures or functionalities to proteins that are far outside of what’s naturally there. Maybe you could get them to do new and better things that are really useful for people for a whole variety of applications, not the least of which would be drug development. That’s what I’m most excited about.
Now that we’ve published this, I’m hoping that other people will attempt to retrieve our increased information in a whole variety of novel ways. Not only for protein synthesis for drugs, which is what I’m going to focus on, but one could imagine trying to evolve bacteria that harbor novel enzymes that help them degrade pollutants or toxins, reduce green-house gases, or produce insulin better or small molecules in a different way.
There’s all sorts of things I could imagine you could use this information for and I’d be really excited. The impact that I hope it has is that people begin to do new and interesting things with the increased information.
Where can readers find more information?
http://www.scripps.edu/romesberg/
About Associate Professor Floyd E. Romesberg
 Dr. Floyd E. Romesberg is currently an Associate Professor of Chemistry at The Scripps Research Institute in La Jolla, California. His research combines the tools of bio/organic chemistry, molecular biology, microbiology, genetics, and modern spectroscopy to study different aspects of evolution.
Dr. Floyd E. Romesberg is currently an Associate Professor of Chemistry at The Scripps Research Institute in La Jolla, California. His research combines the tools of bio/organic chemistry, molecular biology, microbiology, genetics, and modern spectroscopy to study different aspects of evolution.
Projects include the identification and development of novel antibiotics, the development of tools to apply steady-state and time-resolved UV/vis and IR spectroscopy to understand how proteins are evolved for biological function, the investigation of the cellular response to DNA damage in prokaryotic and eukaryotic cells, and the development of unnatural base pairs with which to expand the genetic alphabet and thereby increase the chemical and functional potential of DNA and RNA.
In addition, he is a co-founder of Achaogen, Inc. and RQx, Inc., two companies working to develop novel antibiotics, as well as Synthorx, Inc., a new synthetic biology company.