Sponsored Content by PittconApr 21 2016
Please can you give an introduction to your research and your talk at Pittcon?
In our bioanalytical mass spectrometry lab we use proteomics techniques to try to understand more about Alzheimer's disease. The primary thrust of our research is that we're interested in understanding the changes that take place outside of the brain and how those correlate with what's taking place inside the brain.
We study tissues in the immune system. We study tissues like the liver and other peripheral organs and then correlate how protein-expression changes in those tissues track with the same types of changes that are taking place in the brain. It's a different way of thinking about Alzheimer's disease, outside of the brain.
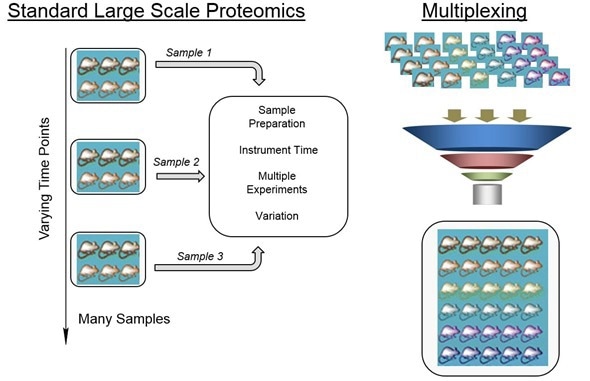
In my talk at Pittcon 2016, I explained some of the challenges that we encounter with processing information from the types of experiments that we carry out. Our data file sizes are ginormous and are hundreds of gigabytes by the time we put them all together. We get a lot of information about hundreds to thousands of proteins. What do you do with that information and how do you take that information and turn it into knowledge about Alzheimer's disease?
Mass Spectrometry in Proteomics
Mass Spectrometry in Proteomics from AZoNetwork on Vimeo.
Why are ‘omics-based approaches being increasingly used to investigate biological processes and, in particular, to increase our understanding of Alzheimer's disease?
‘Omics-based approaches are extremely powerful in understanding the biology of any system. The idea behind ‘omics-based approaches is that you can look at hundreds to thousands of molecules simultaneously in a single experiment and obtain information about how they change.
Traditionally, you might have looked at one protein at a time and it might have taken two years to really understand the expression of that protein using biological assays. However, now you can look at a thousand or maybe two thousand proteins or metabolites at one time, so it really just changes the entire dynamic in terms of how fast you can get through an experiment and actually reach the stage of getting information about disease.
In the last ten years, there's been an increase in the number of groups that have been taking advantage of these techniques in order to understand Alzheimer's disease. These techniques really give us a lot more insight than what we could gain from looking at one protein or metabolite at a time.
What are the main levels with which one can answer biological questions about Alzheimer’s disease?
There's really a big funnel in terms of the levels at which you can start to think about this disease. For our group, we are interested in taking this very basic molecular information about proteins and then connecting that back to some of the changes that take place in the clinic or that take place with memory and seeing how those things eventually correlate.
You can think about levels in terms of what the changes are that take place in the clinic from the point of view of the patient and what the doctor sees, to what some of the clinical techniques are that doctors can use to diagnose someone with Alzheimer's disease.
You can think about changes that take place in terms of populations of people. You can think about what the patterns and similarities are across groups of people, what potential biomarkers there are that exist for the disease, all the way down to what the genes are that are associated with increased risk of disease, what the specific types of proteins are that are changing with disease in those populations, down to metabolites and other types of molecules.
Why is synthesizing information from these multiple levels challenging?
The challenges exist because there aren't quite yet individual software packages that really allow you to do these things. There's just a mix of different software platforms that allow you to look at either proteomics data or genomics data or to look at clinical population data.
To date, there is not one fully integrated program that allows you to pull all that information into one platform for the user. That can become challenging because we find the students end up having to take proteomics information, then look up other people's proteomics information, compare those data and then try to relate that back to the clinic. Getting to that is a very slow process.
How can these challenges be overcome?
The start is really just getting together the people that are looking at disease on different levels so they can start to communicate with each other in order to figure out how we can develop something that would serve all of our needs.
I think the biggest thing is people talking and then getting some dedicated people that will take existing databases and programs that are available and then combine them either into one new database or generate something new that can meet the needs of proteomics people, clinicians and all the people that are really interested in looking at the different levels of Alzheimer's disease research in one place.
How have mass spectrometry and bioinformatics advanced in recent years and what impact has this had on your research?
Mass spectrometry has just taken off in the last ten years or so and even more so in recent years. The types of instruments that we have are just phenomenal in terms of their sensitivity and throughput. The types of experiments that we can run has changed.
What this does for ‘omics analysis is give us better instruments with which we can create data that we're more confident in and we can do that in a much faster way. We can start to answer all types of different fundamental questions about proteins and metabolites that we haven't been able to answer before.
For our research particularly, the growth in mass spectrometry instruments and in the number and types of bioinformatics platforms has really given us more insight about Alzheimer's disease. We benefit tremendously by having very sensitive and fast instrumentation that we can use to carry out proteomics experiments.
What are the main applications of mass spectrometry in this field?
In proteomics, mass spectrometry is one of the most powerful separation and mass analysis and detection systems available. It's used as a device that allows us to separate molecules of different mass-to-charge ratios in the gas phase. From that separation of masses, we can obtain information about structures of proteins, sequences of proteins, interactions of proteins with other molecules such as proteins and small molecules.
We can look at the structures of lipids and of metabolites. We can get a whole plethora of information just by measuring the mass-to-charge ratio of a molecule. It's really the gold standard for doing proteomics experiments today.
Is current technology limiting your research in any way? What do you think the future holds for this field?
Not so much in terms of instrumentation. I think the current limitation is just having the right suite of bioinformatics tools available that allow us to gain knowledge more quickly. We generate a lot of information from our experiments, having the right tools available to help us gain knowledge is probably one of the challenges and a limiting factor.
I think the future is pretty wide open. I think instruments will continue to get better in order to allow you to do more types of analyses and gain more information from biologic systems. I think that the informatics tools are being developed as we speak and will continue to become more available and more widespread.
I really think what will happen in the future is that we will get to a place where we can take a tissue sample, inject it into an LC mass spec system and, within minutes - hopefully no more than hours - we'll gain a lot of useful and direct knowledge about disease. I think we'll be able to do that really quickly in the future.
Which talks at Pittcon 2016 did you find particularly interesting and relevant to your research?
The Emerging Leaders in Biological Mass Spectrometry session that I was a part of was an extremely insightful and well-done session, it was critical for our research because it gave us opportunities to understand how other groups are approaching biological processes.
Listening to the work of Ying Ge, Amanda Hummon, Yu Xia, and Heather Desaire gives us a different way of thinking about how we can approach the problem, even more so than we do now.
What Pittcon Can do for You
What Pittcon Can do for You from AZoNetwork on Vimeo.
It's also been helpful to sit down at some of the talks about instrumentation advancement and to learn about high-resolution mass spectrometry and what the state of the art is with instruments today. For example, hearing about the 21 Tesla Magnet and the state of Orbitrap has been very interesting for us.
What are the key benefits you believe people gain from attending Pittcon? What does Pittcon mean to you?
I think Pittcon is an excellent opportunity for students and for faculty to come and learn about leading-edge research and gain some insights into what analytical groups are doing at the moment – things that may not necessarily be published yet. I think it's a great opportunity for students to be exposed to research and have the opportunity to present their research.
For me, it's a great opportunity to window-shop and see what the tools are that we need to have in the lab and how we can update our instrumentation to make our process a lot more streamlined and more throughput. Pittcon allows me to do all of those different things.
Where can readers find more information?
www.pitt.edu/~rena
About Dr Renã Robinson
Dr. Renã Robinson is currently an Assistant Professor in the Department of Chemistry at the University of Pittsburgh.
She was a Lyman T. Johnson and UNCF/Merck postdoctoral fellow, received the 2010 Society of Analytical Chemists in Pittsburgh Young Investigator Award and the 2014 Lloyd N. Ferguson Young Investigator Award.
She is developing high throughput proteomics methodology to study aging and age-related diseases, such as Alzheimer’s disease.
She has published over 40 peer-reviewed articles and reviews and has authored 4 book chapters.
About Pittcon
 Pittcon® is a registered trademark of The Pittsburgh Conference on Analytical Chemistry and Applied Spectroscopy, a Pennsylvania non-profit organization. Co-sponsored by the Spectroscopy Society of Pittsburgh and the Society for Analytical Chemists of Pittsburgh, Pittcon is the premier annual conference and exposition on laboratory science.
Pittcon® is a registered trademark of The Pittsburgh Conference on Analytical Chemistry and Applied Spectroscopy, a Pennsylvania non-profit organization. Co-sponsored by the Spectroscopy Society of Pittsburgh and the Society for Analytical Chemists of Pittsburgh, Pittcon is the premier annual conference and exposition on laboratory science.
Proceeds from Pittcon fund science education and outreach at all levels, kindergarten through adult. Pittcon donates more than a million dollars a year to provide financial and administrative support for various science outreach activities including science equipment grants, research grants, scholarships and internships for students, awards to teachers and professors, and grants to public science centers, libraries and museums.
Visit pittcon.org for more information.