Mar 7 2017
Researchers at Okayama University report in Scientific Reports how the overloading of protein transport mechanisms affects cell functioning. Over-expression of different types of target proteins leads to cellular growth defects, and the mechanism for transporting proteins out of a cell nucleus has the lowest over-expression tolerance.
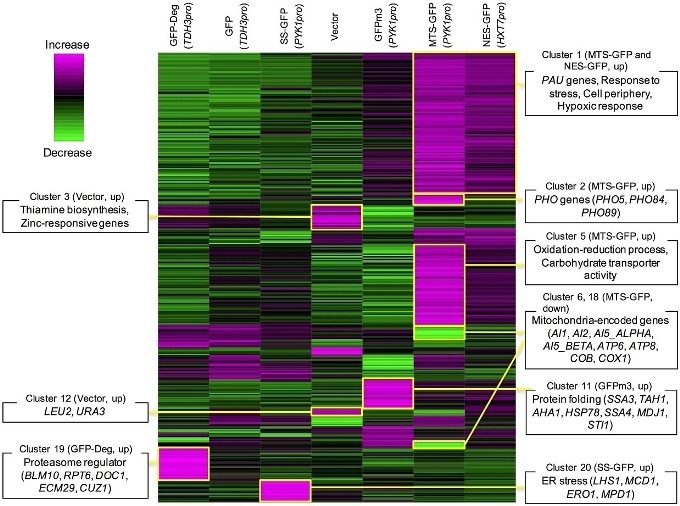
Representation of changes in gene expression levels (green: decreased, violet: increased) upon high-level expression of different types of modified green fluorescent proteins (GFPs). Clusters of genes with specific functional categories are marked by boxes; clearly, high-level expression of modified GFPs leads to defects in cellular functioning.
Transporting proteins to where they need to be within an organism’s cell — protein targeting — costs resources. Too much expression (the synthesis and subsequent handling by the cell) of proteins that require transportation may limit the cell’s resources for moving other proteins around, potentially hampering proper cell functioning. A team of researchers from Okayama University led by Hisao Moriya has now investigated in detail what actually happens when protein-targeting resources become overloaded, and concluded that overloading situations lead to cellular growth defects.
For protein targeting, organisms rely on so-called signals: particular amino acid sequences within proteins that act as labels. Mitochondrial targeting signals (MTSs), signal sequences (SSs), nuclear localization signals (NLSs) and nuclear export signals (NESs) are important labels that drive proteins to mitochondria, to the endoplasmic reticulum, into the cell nucleus, and out of it, respectively. Moriya and colleagues attached signals of these types to green fluorescent proteins (GFPs) and, using a method they had developed before (‘genetic tug of war’), measured the expression limits of the modified GFPs.
By comparing results for signal-modified and unmodified GFPs, the researchers first observed that the added signals reduce expression limits of the modified GFPs — except for the NLS modification. NESs had the lowest limit; this is due to the exhaustion of a protein known as chromosomal maintenance 1 (Crm1) when overloading nuclear export processes mediated by Crm1. For MTS- and SS-modified GFPs, specific limiting factors could not be identified yet.
Moriya and colleagues assessed the physiological consequences of the reduced protein expression capabilities by gene analysis of the sets of messenger-RNA molecules (the transcriptomes) in the cells expressing modified GFP; they found that the artificially applied signals trigger various defects in cellular processes, similar to what happens with GFPs modified with a misfolding mutant.
The scientists also managed to obtain estimates for the critical amount of protein causing growth defects, via measurements of the number of modified GFPs produced. For MTS-, NES- and SS-modified GFPs the limits were 4%, 1% and 0.7%, respectively (the percentages are relative to the amount of unmodified GFP produced).
The findings of Moriya and colleagues show that the overloading of transport machineries in cells has various knock-on effects on cellular functioning. As the researchers point out: “understanding and controlling process overloads is beneficial for disease treatment and cellular engineering.”