Introduction
This article presents a comprehensive genome wide comparison of native and cross linked chromatin immunoprecipitation (ChIP) and next generation sequencing (ChIP-seq) using the patented solid state Chromatrap® spin column technology.
ChIP is a technique used to analyze protein DNA interactions and can be divided into two methods such as Cross linked (X-ChIP) and Native (N-ChIP). Native chromatin prepared by enzymatic digestion is used by N-ChIP, which can be employed to study histone modifications as well as transcription factors that are tightly bound to DNA. XChIP, on the other hand, utilizes chemicals to cross link the protein and DNA components of chromatin, and is suitable for histone modifications as well as more loosely bound, or low abundant, transcription factor protein targets. These two assays can be carried out at locus or gene specific sites through qPCR and can also be used to perform genome wide analysis via downstream sequencing (X or N-ChIP-seq).
Chromatrap® offers high-quality X-ChIP-seq kits (Cat no. 500189 & 500190) and N-ChIP-seq (Cat no. 500237 & 500238). In this article, the enrichment of the histone H3K4me3 modification signal is compared across the entire genome, from chromatin prepared by native vs cross link methods. For this analysis, the histone mark H3K4me3 was selected to demonstrate the efficiency and sensitivity of the Chromatrap® spin column technology for N-ChIP and X-ChIP-seq methods. H3K4me3 is associated with gene activation and ‘open’ transcription start sites. It is an abundant epigenomic target and is present in euchromatin across the genome.
Method
Chromatin preparation
The human endometrial epithelial cancer cell line Hec50 (Holinka et al., 1996) was used to prepare chromatin for X-ChIP and N-ChIP assays according to the Chromatrap® protocols. For the N-ChIP assay, 1 x 106 cells were grown to a confluency of 80%, scraped in ice-cold PBS, and finally collected through centrifugation. Cell lysis was performed in Hypotonic Buffer, before subjecting the separated nuclei to micrococcal nuclease digestion to yield chromatin fragments between 100 and 500 bp in length. Sample dialysis was carried out to remove unwanted impurities and these samples were processed for the ChIP method. For the X-ChIP assay, 1 x 106 cells were grown to a confluency of 80% and fixed in 1% formaldehyde for a period of 10 minutes. Glycine was used to quench the fixation reaction and cells were gathered in ice-cold PBS and then spun down, discarding the supernatant prior to re-suspension and lysis in Hypotonic Buffer. The nuclei thus released were gathered by centrifugation and lysed, and the chromatin was sheared to acquire fragments between 100 and 500 bp in length.
Chromatin IP
Slurries were prepared for immunoprecipitation at a 5:2 chromatin: antibody ratio, 5 μg chromatin: 2 μg of H3K4me3 (Cat no. 70010). Prior to immunoprecipitation with the Chromatrap® spin columns, slurries were incubated for a period of 1 hour at 4°C in accordance with the X-linked or Native protocols. Inputs containing 5 μg chromatin were also prepared separately. These samples were not exposed to ChIP enrichment but used for analyses. After the X-ChIP samples were eluted, they were reverse cross-linked for a period of 2 hours, and this was followed by proteinase K digestion. Then, N-ChIP samples were briefly exposed to proteinase K digestion. Chromatrap® DNA purification columns were used to purify all the samples prior to library preparation.
Library preparation
The NEBNext® Ultra™ (NEB) library preparation kit was used to prepare libraries, and the KAPA Biosystems (Roche) quantification kit for NGS was used to quantify these libraries. Then, an Agilent Bioanalyzer (Agilent Technologies) was used to process the samples in order to verify sample size distributions from 200 to 300 bp, as indicated in Figure 1.
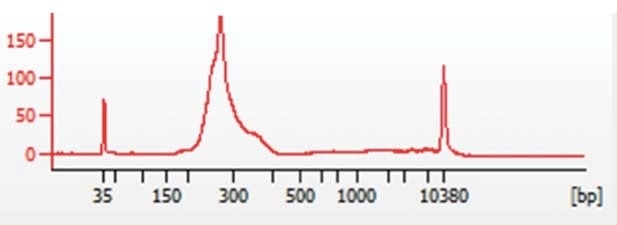
Figure 1. Representative Bioanalyzer trace showing chromatin sample size distribution between 200 and 300bp.
Chromatrap® ChIP-seq analysis
Using the Illumina® MiSeq, paired end (2x75 bp reads) high throughput sequencing was carried out and u the quality of the sequencing data was assessed using FASTQC. The ENCODE UCSC genome browser database was used to map the sequences to the human reference genome (hg19) and Bowtie2 was used to align these sequences. Next, peak enrichment data were examined by MACS2 using Chromatrap® ChIP-seq data analysis software, and input for X-linked and N chromatin samples was provided as baseline/background. Enrichment of H3K4me3 peaks for the X-ChIP and N-ChIP samples was observed using Integrative GENOMICS viewer online software (IgV1.4)
Results
High-quality sequence scores (FASTQC software) were obtained from both X-ChIP and N-ChIP. Duplication rates were less than 5%, and Q scores above 30 were noted for all samples. Q30 scores correlate to a probability of 1 in 1000 of a base being called inaccurately. Figure 2 A-C shows the FastQC scores and the following duplication levels were obtained for H3K4me3 for XChIP and N-ChIP..
A. Sequence quality and duplication
|
Sample
|
Q30
|
Duplication rates
|
|
N-ChIP H3K4me3
|
>30
|
4.13%
|
|
X-ChIP H3K4me3
|
>30
|
2.58%
|
B. Sequence duplication levels for N-ChIP and X-ChIP respectively
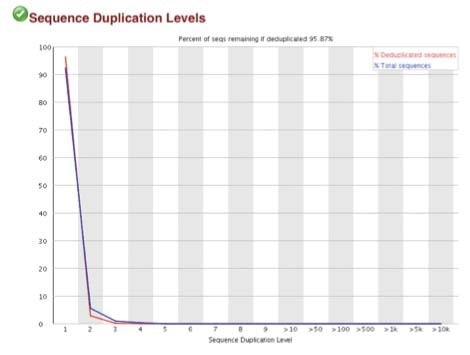
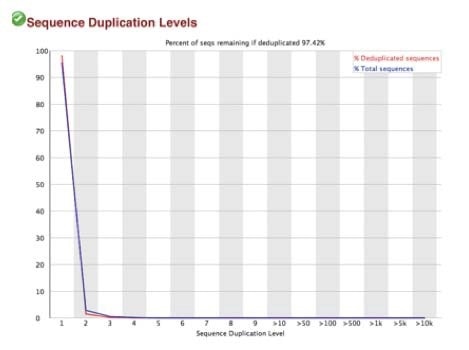
C. Q30 scores for N-ChIP and X-ChIP samples respectively
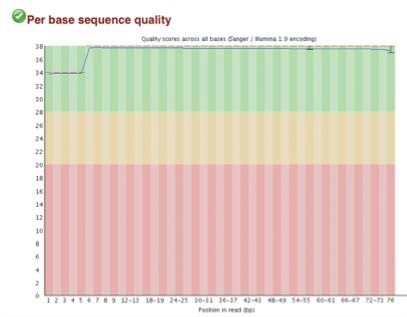
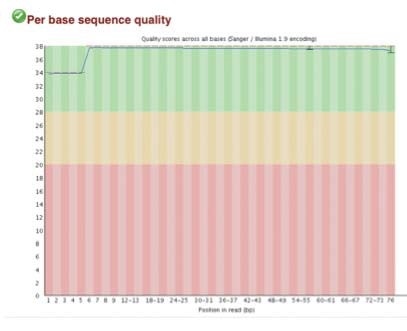
Figure 2. FastQC sequence quality and duplication for both N-ChIP and X-ChIP sequenced samples.
Chromatrap® ChIP-seq Data Analysis software was used to analyze X-linked and Native sequenced samples for peak enrichment. For N-ChIP samples, strong enrichment was observed for 65,000 peaks, ranging from 3 to more than 30 times enrichment over the input. 39,000 peaks with strong enrichment were obtained for the X-ChIP assay..A higher number of peaks may have been obtained from N-ChIP due to the possibility of formaldehyde cross links masking protein epitopes, making them less accessible to the antibody.
For both X-linked and Native chromatin samples, low resolution genome wide IGV visualized peak enrichment revealed similar peaks for H3K4me3. Figure 3A and 3B show the peak maps, indicating equivalent levels of H3K4me3 enrichment across the genome for N-ChIP and X-ChIP samples, respectively. This shows that on a genome wide scale, Chromatrap® immunoprecipitation is comparable and reproducible between X-ChIP and N-ChIP-seq.
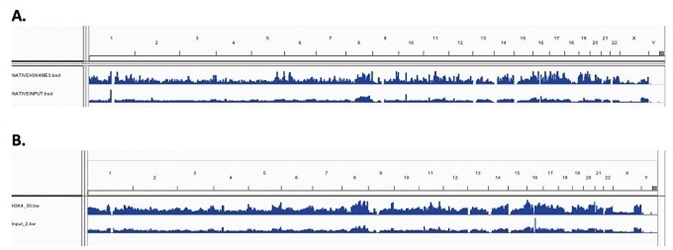
Figure 3. Enrichment peak maps of H3K4me3 across the whole genome (A) N-ChIP (B) X-ChIP
Uniquely identified genes
In order to determine the similarities in the H3K4me3 genome wide landscape, the identified peaks were
mapped to their closest gene loci (+/-5000 bp distance to the TSS to establish which peaks correlate to each gene), and the uniquely identified genes were compared between both X-linked and N-ChIP samples. As shown in Figure 4, 20,315 uniquely mapped genes and 19,508 unique gene loci were shown to be associated with H3K4me3 in the N-ChIP samples and H3K4me3 in the X-linked ChIP samples, indicating as high as 90% resemblance between the samples.
When studying specific gene loci, additional validation of the homology between X-ChIP and N-ChIP-seq is seen. ZNF566, the ubiquitously expressed DNA binding protein, exhibited similar enrichment of H3K4me3 peaks above the background in X-linked and N-ChIP samples (Figure 5).
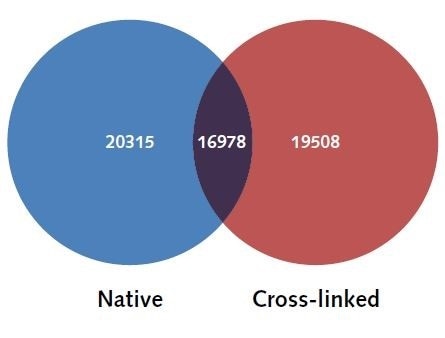
Figure 4. H3K4me3 uniquely identified genes
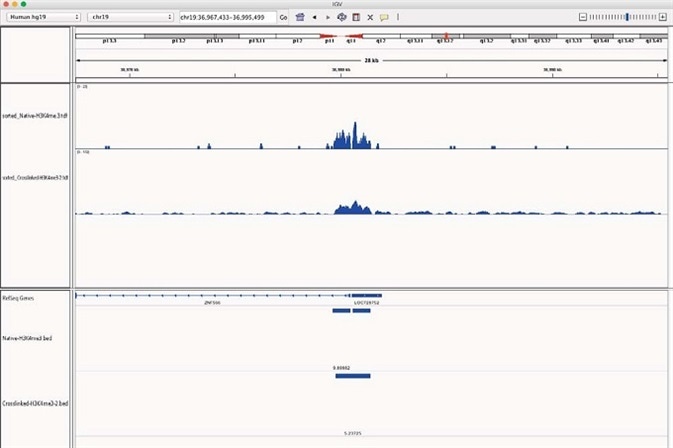
Figure 5. H3K4me3 enrichment onto ZNF566 for both N and X-linked Chromatrap ChIP-seq
Conclusion
This article has shown how Native and Cross-linked chromatin preparation methods allow strong and comparable enrichment of H3K4me3. The work also underscores the fact that both these preparation methods can be used with Chromatrap® extraction and capture technology. Both Native and Cross-linked assays provide excellent data, ensuring better enrichment of H3K4me3 followed by high throughput sequencing. The use of both assay technologies makes it possible to acquire substantial genome wide peak enrichment levels as well as high-quality FASTQC metrics.
Chromatrap® provides an extensive range of ChIP assays that offer users an abundant choice and flexibility to perform different chromatin preparation and enrichment methods for downstream next-generation sequencing.
About Chromatrap®
 Chromatrap® is a product of Porvair Sciences, a wholly owned subsidiary of Porvair plc. We are one of the largest manufacturers of Ultra-Clean microplates, 96 well well filtration plates and Microplate handling equipment for life science and synthetic chemistry. With offices and Class VIII clean room manufacturing located in the UK, combined with a world-wide network of distributors and dedicated distribution hub in the USA, we pride ourselves on our continuous innovation, research and flexibility to meet customer demands. We offer OEM production and contract manufacturing through our North Wales facility.
Chromatrap® is a product of Porvair Sciences, a wholly owned subsidiary of Porvair plc. We are one of the largest manufacturers of Ultra-Clean microplates, 96 well well filtration plates and Microplate handling equipment for life science and synthetic chemistry. With offices and Class VIII clean room manufacturing located in the UK, combined with a world-wide network of distributors and dedicated distribution hub in the USA, we pride ourselves on our continuous innovation, research and flexibility to meet customer demands. We offer OEM production and contract manufacturing through our North Wales facility.
Our porous polymeric material, BioVyon™, whose chemical functionalisation can endow it with internal surface properties individually configured to capture and separate target species out of difficult mixtures, has opened up many possibilities in the field of BioSciences where molecules of interest such as DNA, RNA, proteins etc can be selectively pulled out of complex mixtures of biological origin. The materials have proven to be a remarkably good substrate for accepting novel chemistries such as the organically bound Protein A and Protein G in Chromatrap®.
Using our 25 years experience of microplate manufacturing, Porvair Sciences has now developed a high-throughput bead-free ChIP assay based on our filtration plates containing our Chromatrap chemistry. Chromatrap-96 enables large scale epigenetic screening to become a reality in many laboratories and eliminates many of the long and laborious steps previously undertaken in such work.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.