What first attracted you to the field of MPI?
In around 2004, there was a Phillips paper that discussed a new imaging technique called MPI. At that time, I had an eager, promising graduate student named Matt Ferguson who wanted a project, so I asked him to take a look.
In the paper they seemed to be limited by the tracers; they were claiming that the tracers were giving them only 3% signal.
They had used these commercial tracers and only 3% were the right size. I told Matt we could definitely do better than that because, not only can we make the tracers the right size, but we can also do the modeling, figure out what size really works and then make the size that we want.
Matt started his thesis with that in mind and the very first tracers that we made after we did the modeling gave a 90% signal rather than the 3% that Phillips reported in the first paper.
Magnetic Particle Imaging | Innovation with Simple Physics
How did you get involved in MPI and biomedicine?
I come from a very strong background in magnetism and magnetic materials, so I'm broadly interested in magnetic materials. However, one of the reasons I moved here from Berkeley in around 2001 was that I felt that there was a big opportunity for applying magnetic materials in biomedicine, in both biology and in medicine.
I always like to give credit to my students and I started with a student whose name is Marcela Gonzales. She was a Ph.D. student here and she and I started to learn biology together in order to apply magnetic particles to biology.
We started from the biology side and, over the years, we have largely focused on the applications of magnetic nanoparticles in biology. The first thesis that Marcela did was applying magnetic particles to deliver local heat and applying an AC magnetic field to kill cancer cells. Then, Matt came along and we applied the knowledge that we had to develop tracers for MPI.
Then, I had another student, Ahmed, who tried to combine the two works together and develop a particle that could be jointly applied for imaging and therapy, so hyperthermia and MPI. Now, I have another student who’s just finished, Ahmed Aramhi, who is going as a post doc to Stanford. He started to move all of this towards in vivo experiments, animal models.
What current research topics are you working on with particles? What are you particularly excited about in this field at the moment?
In some ways, we are at a very, very exciting point in our research. We have all this broad knowledge across magnetism, magnetic materials, surface functionalization, some in vitro with cells and what I find really exciting is that we are poised to do some incredible pre-clinical work in vivo at this point.
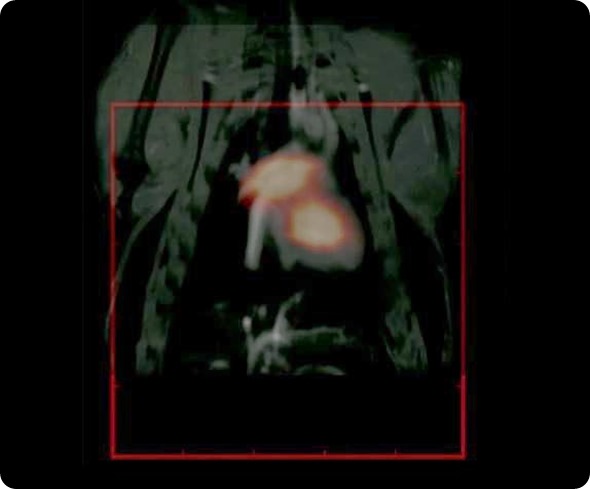
We are really ready to look at how this can be applied, for example, in molecular imaging for cancer and perhaps cell tracking, stem cell tracking and cardiovascular imaging for circulation of the particles.
I think we have incredible opportunities right now because everything has reached a point where we can do a variety of what I think are rather interesting pre-clinical biomedical applications in vivo.
Is the field at a point where it's ready to go to biologists?
This is quite a challenge. For the magnetic particles and especially the magnet-type particles, the chemistry and how the particles are made is well known. What is not so well appreciated is that for MPI, you need particles of a very well defined size with a nano-sized distribution.
People understand that now, but in addition to that, you also need very high phase purity because it's sort of a mixed-valent state of iron in the magnetite. You do not get complete phase purity if you're not careful. That is one of the things that we've been emphasizing – size, size distribution and phase purity - that defines the core and is the first part.
The second part is what you put on the surface. Even if you have the core, different people have different chemistry roots to get the surface functionalized. We are trying to make a well-defined platform and other people have their own platforms.
Having said that, what is really limiting and holding people back is that individuals who are not directly on board with MPI or MPS have not really seen what MPI can do. A lot of it really depends on us promoting it and telling people what MPIs strengths are.
MPI has two important strengths. Firstly, the tracer that we're using is quite biocompatible and it has already been approved for many things. For the clinical part, it's an approved tracer and can be easily approved.
The second point, which is really important, is that in MPI, the signal is completely linear with concentrations. You can actually quantify these images.
A third point is that the surface chemistry is not that complicated. That’s the area that we are now really focusing on. For example, we have recently functionalized for circulation time and we have shown 160 or 180 minutes of circulation in mice, which is quite large because the heart beats at 420 beats per minute, meaning that, for humans that could be a factor of 6 or something larger, which is a very large circulation time. We can also functionalize them for specific targeting, which would mean they would be taken up by specific cells and so on.
All of these things together mean we're at the point where we can promote MPI and magnetic particle spectroscopy (MPS), especially at the pre-clinical level, to a much larger audience across multiple areas of biological and medical research.
How do you make a nanoparticle? Could you give a summary of the process?
People have been making nanoparticles for a very long time. It's not something new. What is really important for MPI, is that the particle has to be a given size in a very narrow size distribution.
What people have been doing traditionally is making particles in water- they come out quite nicely, but they have a very large size distribution. These were the original particles that Phillips tried and they had a very large size distribution.
We use organic solvents, specifically a very traditional method called LaMer synthesis, where you take a precursor that will decompose when you heat it to a certain temperature. You provide the right conditions so that when it decomposes, they can agglomerate and form a particle at those reaction conditions.
In this process, we can control this very accurately, by changing the ratio of the precursor to the other chemicals in there such as the surfactants. By controlling those ratios, we can control the size. Then, the whole reaction occurs in such a way that, once we control the size of the particles, they all literally nucleate at around the same time so that they all grow to the same size.
You control the ratio that controls the size and then, in time, you have a very discrete nucleation event where all the particles nucleate at a very discrete point in time and then they all grow to almost the same size.
In order to achieve this, we have to do it in organic solvents. The big challenge here once you're in the organic solvent, with the right size and the narrow size distribution, is transferring it to water. Then you can use it for biological applications.
We have very nice ways of performing the transferal process. What we now do is use the molecules that we have, the polymers on the surface that we use to do the transfer. We use those to functionalize for different targeting and so on. That's the very simple idea in a nutshell.
You're very heavily equipped for a lot of different measurements but when you're in quality control, what are the critical measurements you put in place to make sure you're getting what you want?
First, we did a lot of modeling to see what the optimal size of a particle for a 25 kHz imaging system is. Our modeling takes into consideration how the particles behave when you have an alternating field that changes with time on a very short timescale and based on that, we have generally come to the conclusion that the optimal particle size is somewhere between 24 and 27nm.
What our quality control then entails is how to make sure that the particles are between 23 and 27nm. That's the first question, the core size. So, we make a number of measurements. We take the particles, we measure their magnetic behavior and then we can do some modeling of the magnetic behavior that will give us the core size from the magnetic measurements.
We also measure physically; we image these particles by TEM. We measure large numbers, maybe 10,000 particles and we also measure the size. These are the two measurements because the magnetic measurement measures much larger quantities, so it's much more statistically significant. Then we look at the visual part of it using the TEM, 10,000 particles. That's how we establish the core size and the size distribution.
We also have another requirement coming from the biology, which is what the hydrodynamic size of the particle will be. In order to get circulation and prevent it from being absorbed in the kidney, it has to be around a certain hydrodynamic size. Approximately 10 to 15nm is the size where the particle is not absorbed by the kidney.
Already the core size is 25nm, so we don't have to worry too much. Our particles will not go into the kidneys, but there is another size, which is the upper limit. That is determined by the spleen and so on for maximum circulation and that size is about 100 to 120nm. We tried to make the hydrodynamic size about 60nm, which would optimize the circulation time.
We then have to measure the hydrodynamic size, which we do using dynamic light scattering. In our set-up, we actually have the particles and we measure the hydrodynamic size. We have to measure not only the hydrodynamic size, but we also have hydrodynamic size in the organic solvent first. Then, we transfer it to the water phase and look at that.
When you come from a physics background you just stop at that point, but that’s not where we stop. If you look at the biology and what happens to these particles in the body, there are proteins that absorb onto the surface called opsonins and they opsonize the particle. So, we actually now measure all of this in biological media where there is a cocktail of proteins and we see how the hydrodynamic size changes.
Once we are sure about the hydrodynamic size and the core size, we move to the magnetic particle spectrometer. We measure the particle response in the organic medium, in the water solution and then in the biological medium, because if the particles are opsonized and they aggregate, then the performance will be deteriorated. Therefore, we actually measure them there.
After that, if possible, we will internalize or work with cell cultures to see what happens to the performance of the particles as they go. There are different levels of optimization as we move towards biological applications, especially in the pre-clinical or clinical applications.
Can you talk about some of the recent successes you've had with taking cells and starting in vivo experiments?
We have now done three or four very important in vivo experiments. We don't actually have an MPI imaging system, so we have to collaborate with people who have MPI systems. I'm very thankful that we have a very active collaboration with Steve Conolly at Berkeley where we do some of the MPI imaging with his pre-clinical system.
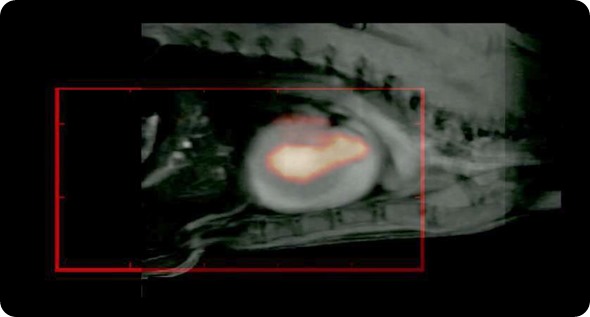
We also collaborate with people at UKE, where they have a Bruker system. We collaborate with the whole group, Michael Cowell and various other people in UKE. We have also started a collaboration with Volk Machules at Aachen. He has the first MPI, the pre-clinical system that was built at Phillips and he's going to get one of the newer Bruker machines. We also collaborate with the people at Wurzburg who have this travelling wave MPI.
Due to the geography, most of our really active collaboration is with Steve Conolly because he is the closest one to us. Of course, we are still separated by 800 miles, meaning we can't do it on a regular basis, so we use alternative modalities for some of the checks.
With Steve, we have performed some fantastic imaging that is of relevance to cardiovascular imaging. We have worked with rat models. His imaging technique takes much longer to image; it takes about 10 minutes to get a good image, but because our particles circulate for long enough, we have been able to resolve the jugular vein and various other vasculature in the head of rodent models. It is really very spectacular work that we have done with Steve.
In addition to that, we are trying to develop various pre-clinical applications and experiments. What we normally do is use some sort of imaging tracer such as a near-infrared fluorescent tag or radionuclide, so that it allows us to use the available existing modalities to work with our particles.
For example, we have been working with a mouse cancer model of a brain tumor, which is a xenograft model, where the MPI tracer has been functionalized with a near-infrared fluorescent tag. We can inject them and see that it goes into the tumor, which we can image with the fluorescent tag. Where the potential of MPI comes in, is that we can resect these tumors and then take them to the MPS (magnetic particle spectrometer) and measure the signal from these tracers.
That’s the true advantage of MPI: the signal can be quantified in terms of the number of particles that are going into the tumor and how successful our targeting is. We use that kind of combination. Similarly, we combine our tracers with a radionuclide and then use existing modalities such as CT, PET to look at biodistribution. This is very important because, ultimately, what we are really interested in once you put the tracer in the body, is where it is going. Once we know that, the next question is really the long term biological fate of the particles. That is a really open question at this time.
Please can you explain how MPI actually works?
The physics of how MPI works is really very simple. You basically have a magnetic particle, which you excite using an AC field. When you apply an AC field to a magnetic particle, the magnetization of the particle changes with time. When that happens, if you put a small pick-up coil around it, it basically changes the magnetic induction that the pick-up coil will see.
If you have a rate of change of induction in the area of the pick-up coil, it will generate the voltage of the pick-up coil. That is the basic idea of MPI. You have the magnetization, which changes with time; it changes the local induction and the local induction induces a change of voltage in the coil, which you pick up - that is your signal.
What MPI requires is not just a signal but also a way to localize the signal, i.e. where the signal is coming from. That is the key point, which of course is the genius of the people who invented it at Phillips.
The idea is that you apply a field gradient and you have a point of zero field in this two dimensional area. The physics says that when you have a point of zero field, the signal is very high and when you have a high enough field where the magnetic particle saturates, where all the moments of the particle are aligned along the magnetic field direction, then you have a very poor signal. There's very little change with time as you apply the field in that case.
Whenever the particles come to the zero field point, you get a very high signal, whereas you don’t everywhere else. That's how you localize the MPI signal. Essentially MPI is just a simple coil that picks up the signal based on the rate of change of induction.
How do you compare MPI to other techniques?
I think it is a rather interesting technique that could be compared with certain existing modalities. First of all, it's a tracer-based technique, so you could compare it with other tracer-based techniques, which would be PET, SPECT or something where a tracer is being injected.
If you compare it with a PET or SPECT tracer, what are its advantages or disadvantages?
What we see is that this is a non-radioactive tracer so immediately there are significant advantages. You don't need to have the specific extra handling and you don't need people trained in radioactive materials.
If you were going to ask me where it competes, then I would immediately say with PET and SPECT, provided it can provide the same resolution as PET and SPECT. That's the challenge for MPI, to get to that point of resolution where it will be comparable to PET and SPECT when using a non-radioactive tracer.
The second thing is how it compares in terms of intensity, sensitivity. I think MPI has a much higher sensitivity than SPECT. We have to find a balance between sensitivity and resolution. Then, you would start to compare with other existing modalities such as optical modalities. That one is a no brainer because, as everybody knows, optical signals are attenuated in human tissue of the order of 10s of or maybe 100s of microns. Magnetic signals actually penetrate the body and they're not attenuated.
Another important point about comparing with optical methods, is that there are many problems with those methods in terms of fluorescence and signals that are, in some ways, washed away, to put it in very simple language.
In MPI, you have a particle that is giving a signal and there is a diamagnetic background that gives you very little signal. In fact, it is an almost negligible signal. If you look at the signal from the particle, divided by the signal from the background that you contrast and your contrast can be quite, almost, infinite because the background is zero signal, which is a diamagnetic material. All human tissue is diamagnetic.
Compared with PET and SPECT, there are some distinct advantages. There are some advantages, of course, with optical imaging and now we come to the more established methods such as MRI.
It's a challenge to see how MPI could potentially compete with MRI. MRI has its strengths and it's a very established method, it's in every respectable clinic. The real question is what are the advantages and disadvantages?
The one thing that I immediately think of with MRI, is that because we are looking at the relaxation of the protons in the body, which have a very, very weak signal, the magnetic field that you have to apply has to have very high purity. Therefore, you require a very significant investment in the building of the magnetic components of the MRI system.
On the other hand, MPI is looking at the relaxation of particles, which have a billion times more magnetic signal than the protons. Therefore, that translates into a little bit more freedom and flexibility in terms of building your magnetic components. You don't have to have such high magnetic field purity, you have much more give in the system.
Secondly, the fields and the field gradients that you apply, the field amplitudes, are much smaller than what you would apply for MRI. I think, overall, the significant compromise or the advantage of MPI could be much lower cost, provided we can demonstrate the competitiveness of the technique with respect to MRI.
Is MPI like PET in that it needs an anatomical reference?
Correct, that's where the complementarity comes in; you will need an anatomical reference. You'll either need MRI or CT so that you can get the background, where it is. It depends on the application, of course. If you want to make it clinically relevant, you will need an anatomical reference. An MPI could be complimentary with either MRI or with CT.
How does MPI compare in terms of speed?
MPI is almost real-time. There is an issue right now, however. When I've used the existing MPI systems, even though the data is acquired in real-time, the image processing software has not reached the point where the data is seen almost immediately. It takes much longer, but the data acquisition is in real-time.
Given the speed, do you think MPI could get closer to measuring biological processes more accurately?
It's possible. As MPI becomes more popular, this is the sort of thing that people who have the right background and interests in biological processes will start getting engaged in. A lot can be done with the magnetic particle spectrometer.
We have just published and submitted to PNAS a paper that actually uses the spectrometer to detect certain enzymes called proteases that are expressed by cancer cells. We can build certain particle structures that have some linker peptides that can be cleaved only by specific proteases. By using those sort of linker peptides with the particle and understanding the relaxation, we can sense the protease expression outside.
I think there is a huge amount of potential in terms of imaging but a huge area remains unexplored in terms of the diagnostics that people haven't started thinking about. We have started to think about diagnostics where the hardware is not going to be so elaborate; it's just a spectrometer rather than an imaging system.
To answer your question in terms of the biological processes, there are lots of possibilities, but I think people have to figure out where the areas are.
What excites you about where this field will be going?
There is great potential for MPI. I think it will take collective effort from a number of people. There are four groups of people that need to come together to really make MPI reach its potential in a number of different areas. They are the hardware people who build the machines; the people like me who build the tracers and understand the physics of relaxation and the surface functionalization for targeting; the physicists and chemists and then you have the people who do the image processing.
I think there's also a lot of work that needs to be done in terms of image reconstruction and how to get the tool into the hands of a radiologist. They could have a turnkey system that means they can just put something in and then have the image, so that they don't have to wrestle with the whole imaging process right now.
Then the fourth group is the biologists and radiologists who can bring all of this into sharp focus and address the kind of problems that need to be addressed, the critical problems in biology and medicine.
All these four groups of people need to come together. Once we do that, in my view, there's really no limit to what we can do. There's great potential for the technique, but I think there's a lot of work to be done right now.
We are bringing people together from these four different perspectives to work together. We are all trying to work together with people of different expertise. I don't think any single one of these groups can move this forward. You need all four to work together.
Another very important point about making particles is that most people make particles in small quantities. They make particles in 10s of mg or 100 mg quantities. When you start to scale up the synthesis, it's a completely different game and many more things can go wrong with the whole synthesis.
If you want to start making gram quantities of particles so that you can use it for different experiments, then you really run into problems because things don't scale in a way that you expect. For example, if you make even this quantity of particle, this is really the tracer, you will see that it does not behave in the same way as when you make a small quantity of tracer.
There is a real challenge in terms of the particle synthesis because the whole particle synthesis is a kinetically driven process. It is not a thermodynamically stable process. You are really performing a non-equilibrium synthesis process and therefore things can go wrong all the time, especially as you scale. It can go even more wrong than when you make them in small quantities.
People may show very good results with small quantities of particles, but when they start making larger quantities, it simply doesn't do the same thing. That's another very big challenge in terms of making these tracers.
About Bruker
 Bruker is market leader in analytical magnetic resonance instruments including NMR, EPR and preclinical magnetic resonance imaging (MRI). Bruker's product portfolio in the field of magnetic resonance includes NMR, preclinical MRI ,EPR and Time-Domain (TD) NMR. In addition.
Bruker is market leader in analytical magnetic resonance instruments including NMR, EPR and preclinical magnetic resonance imaging (MRI). Bruker's product portfolio in the field of magnetic resonance includes NMR, preclinical MRI ,EPR and Time-Domain (TD) NMR. In addition.
Bruker delivers the world's most comprehensive range of research tools enabling life science, materials science, analytical chemistry, process control and clinical research. Bruker is also the leading superconductor magnet and ultra high field magnet manufacturer for NMR and MRI solutions.