 Analysis of immunohistochemical markers
Analysis of immunohistochemical markers
HistoQuest provides three analysis modes:
- Cell-based (nucleus, cytoplasm, membrane)
- Stained areas
- Small dots (e.g. CISH)
- Skeleton algorithm for membrane staining
Context-based analysis can be done by manually drawing regions of interest. TMA can be analysed and also detected on imported files.
 Analysis profiles can be created and used for automated analysis.
Analysis profiles can be created and used for automated analysis.
HistoQuest is integrated into the following TG scanning & analysis systems:
- TissueFAXS Histo
- TissueFAXS PLUS
- TissueFAXS 120
- TissueFAXS 120 PLUS
- TissueFAXS i Histo
- TissueFAXS i PLUS
- TissueFAXS Confocal PLUS
 HistoQuest is also available as a standalone software.
HistoQuest is also available as a standalone software.
 HistoQuest has importers for the following digital image or slide formats:
HistoQuest has importers for the following digital image or slide formats:
- 3D Histech (.mrxs)
- Hamamatsu (.ndpi)
- Zeiss Axio Scan (.czi)
- Leica (.scn)
- Aperio (.svs)
- Images (.jpeg, .tif, .bmp, .png)
New HistoQuest 6.0 - the easy solution for brightfield analysis
HistoQuest is a brightfield image analysis software for the FACS-like analysis of samples stained with immunohistochemical or histochemical stains.
New features in HistoQuest 6.0
HistoQuest 6.0 comes with new features which improve handling of large samples/number of samples in one project as well as increase the softwares speed:
Full 64 bit support will greatly increase the number of samples which can be analysed in one project.
The latest processing libraries have been integrated which will increase processing time by up to 40%.
The cache from TissueFAXS projects will be used, this eliminates wait time while the project is loaded into the viewer.
Finally, HistoQuest has a heatmap feature in scattergrams.
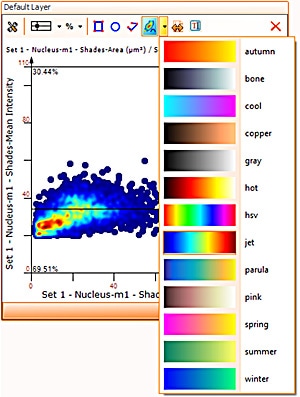
This feature will show areas of objects density in the scattergram which may not be visible in highly populated scattergrams and so deliver more information when working with them.
HistoQuest automation
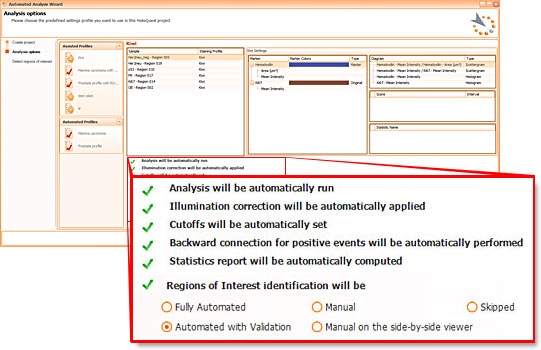
 HistoQuests fully automated template based analysis is optimised for clinical routine and research applications and allows complete walk away operation.
HistoQuests fully automated template based analysis is optimised for clinical routine and research applications and allows complete walk away operation.
The user just has to load samples into the software and to decide whether he wants samples automatically detected (in which case analysis will start immediately), whether he wants to be able to interfere with automatic region detection (with validation) or whether he wants to manually define regions to be analysed.
TGs hidden power - Tissue Cytometry technology
TissueGnostics introduced Tissue Cytometry technology into image analysis software more than a decade ago. While, at a first look, it may appear to be just another form of displaying data, there is much more to it.
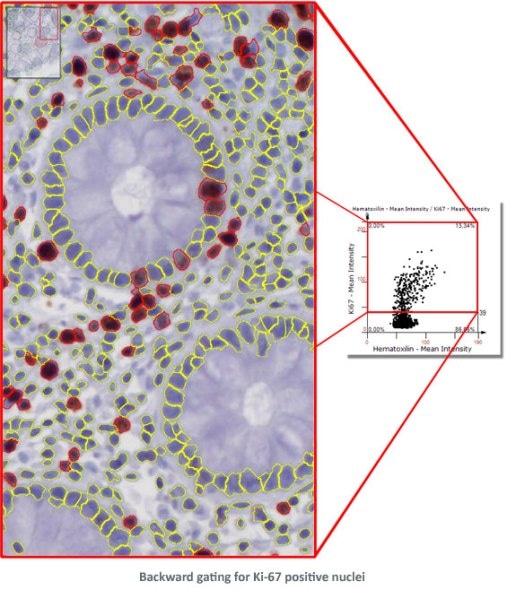
Combined with segmentation images it is also a simple and intuitive tool to visually validate results. TG calls this "Backward Gating".
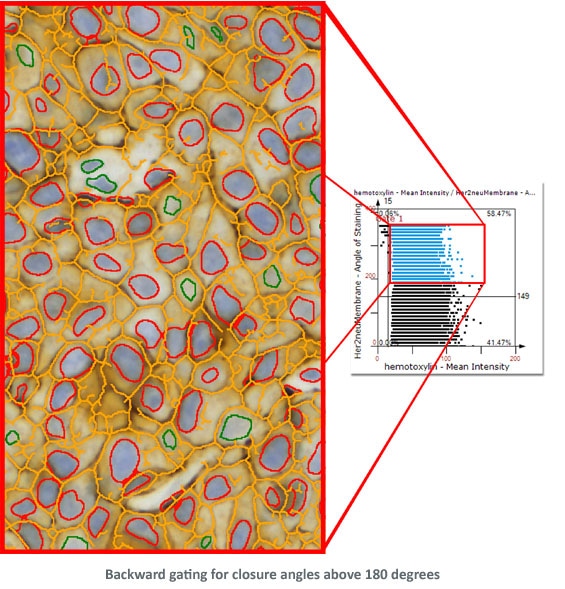
The following examples show Backward Gating versatility on segmented nuclei positive for Ki-67. Backward gating can be done for any measured parameter on histograms and scattergrams.
In the second example backward gating is used to visualise cells in which the Her-2NEU staining, detected with the skeleton algorithm, has a closure angle of more than 180 degrees.
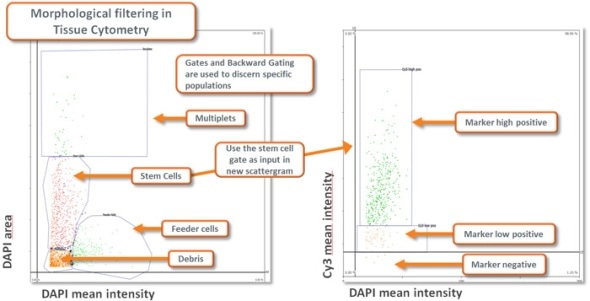
Morphological Filtering using Tissue Cytometry technology combined with appropriate planning of the analysis provides a highly flexible and capable instrument for extracting specific data from a sample (see image below).
HistoQuest Image Analysis algorithms
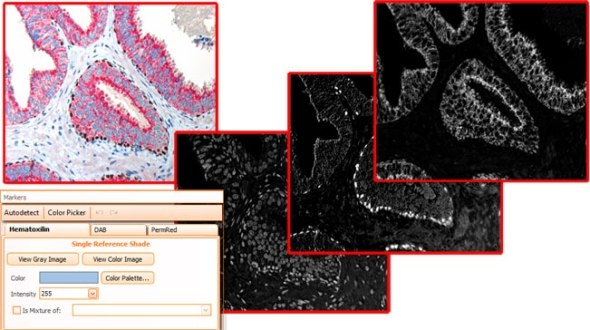
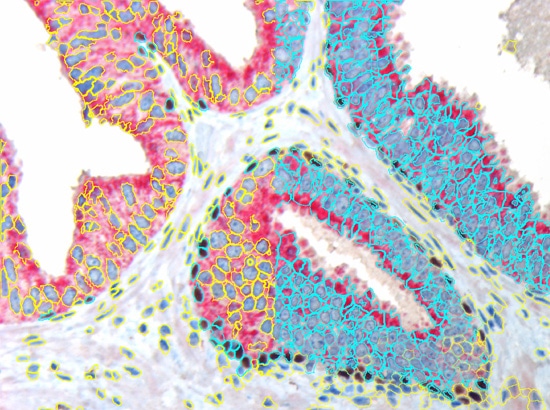
As brightfield images need to separated into images according to their colour components, HistoQuest provides two algorithms for color separation.
The first one automatically detects and separates two colors and can have a third one added (see below).
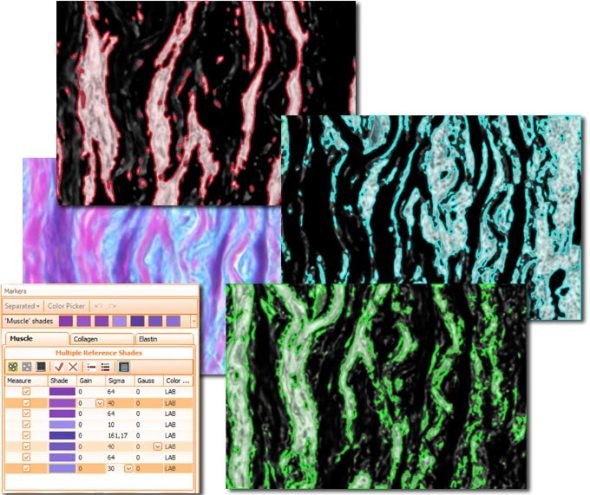
The second color separation algorithm is semi-manual (using a color picker) and separates an unlimited amount of colors with great control and precision (see image with muscle, collagen and elastin detection below).
HistoQuest uses algorithms for nuclear and cell compartment segmentation. It also provides an algorithm for stained area detection as well as algorithms for the detection of dots inside of already detected cell compartments.
The nuclear segmentation algorithm is highly efficient in difficult situations in tissue and can be set with two values only - mean nuclear size and nuclear channel background threhshold. The cell compartment algorithm provides a large amount of flexibility with a few simple settings (see below, yellow=segmented nuclei, blue nuclei positive for DAB or cytoplasm positive for Permanent Red marker).
The Total Area algorithm segments stained areas in a sample (see Muscle/Collagen/Elastin image above).
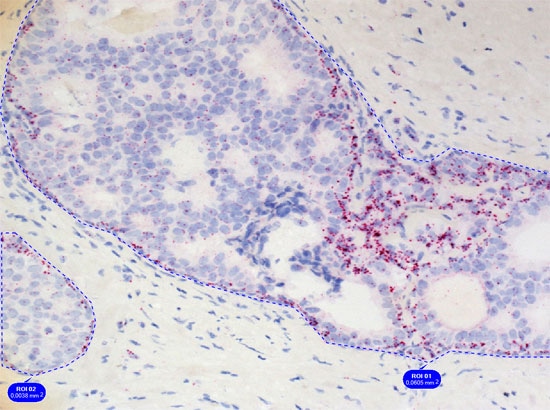
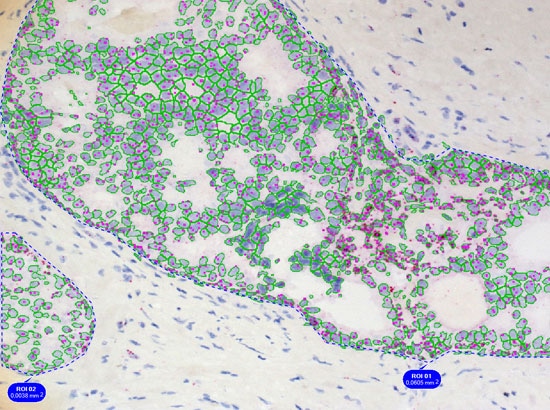
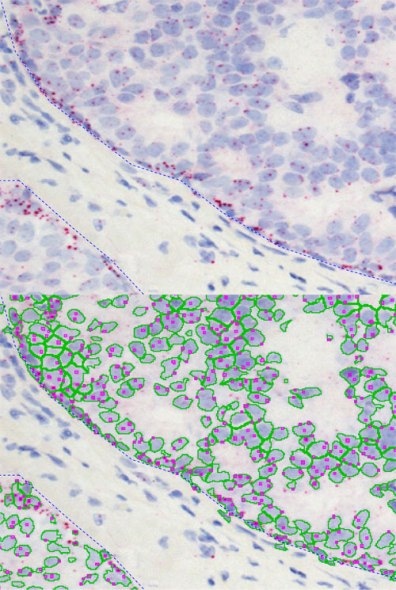
The dot algorithm provides small dot detection ideal for CISH, ISH and other applications in which the quantification of small objects within cellular compartments is needed.
The images below show the quantification of EGFR RNA dots inside of the nuclear compartment.
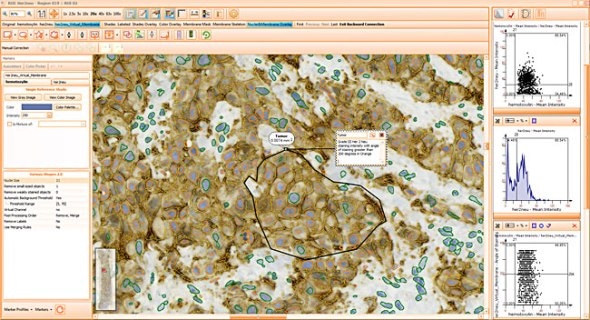
Simple context based analysis can easily be done in HistoQuest by using manual region tools. Drawing the regions takes a negligible amount of time and provides data for epithelial areas and stroma separatelyin this example.
The skeleton algorithm permits the detection of membrane stainings (see image above).