This article and associated images are based on a poster originally authored by Marina Fedorova and Ian Hiles and presented at ELRIG Drug Discovery 2024 in affiliation with GSK.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Background
Drug Discovery faces a rising number of projects that demand the generation of cellular reagents with controlled or lower target protein expression. Here, we have tested and optimized several techniques that can be used to tune protein expression: the inducible system, the stop codon suppression method, the addition of IRES elements prior to the target sequence, and a panel of weak promoters and construct modifications. These methods for customizing protein expression can be combined and applied widely.
Physiological levels of target proteins are required for competitive binding in antibody screening or functional assays. Likewise, matching the surface expression of orthologues is essential to test antibody specificity. Moreover, some targets are toxic when overexpressed; hence, inducible expression is required. Multispecific antibody projects depend upon precise co-expression of several targets; therefore, a combination of methods must be developed.
Program request
- CHO GSKO cells co-expression of Target X and Target Y
- Required to enable screening of the bispecific outputs
- Closely matched to the endogenous expression on primary cells:
- Protein X:Protein Y in a ratio 1:3
- 30-60 K Protein X copies and 10-20 K Protein X copies per cell
Starting point
Method: co-transfection with mRNA transposase
Stable pools & FACS clones: too high expression
- Protein X: 0.1-4x106 receptors
- Protein Y: ~0.1x106 receptors
Objectives
- CHO Protein X: Protein Y tune down 0.1-4x106 to 1-6 x 104 receptors on the cell surface (in the ratio 1:3)
- Develop and optimize methods for precise target expression
Tet-ON inducible system
- The Tet-On transactivator (rtTA) binds to and activates expression from tetracycline-inducible promoter (PTRE) in the presence of Doxycycline (Dox).
- Gene of interest (GOI) transcription and expression should be induced in a dose-dependent manner by Doxycycline
- GOI: Protein X in second position downstream the selection marker Glutamine Synthetase (GS) and the internal ribosome entry site (IRES)
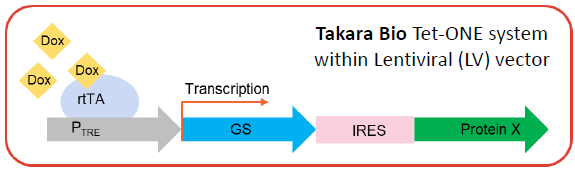
Image Credit: Image courtesy of Marina Fedorova and Ian Hiles in partnership with ELRIG (UK) Ltd.
Conditions:
- CHO GSKO cells
- Lentivirus transduction
- Protein X expression: flow cytometry
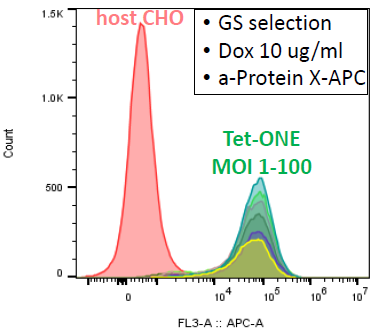
Image Credit: Image courtesy of Marina Fedorova and Ian Hiles in partnership with ELRIG (UK) Ltd.
Under constant Dox expression and GS selection
- All pools express ~ 13-16 k Protein X receptors on the surface
- BUT: cell viability is ~30-40 %
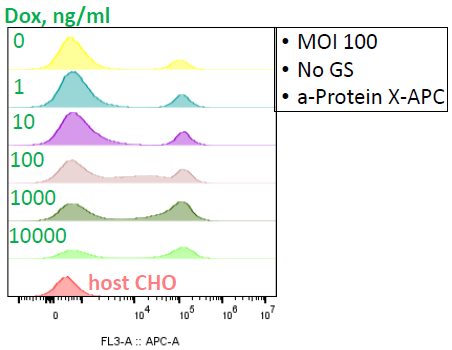
Image Credit: Image courtesy of Marina Fedorova and Ian Hiles in partnership with ELRIG (UK) Ltd.
GS selection is removed, and cells are stimulated with Dox (1-10000 ng/ml) for 48 hours.
- Viability has recovered
- BUT: the presence of non-expressing population
Learnings
- Required Protein X levels achieved
- Non-expressing peaks in pools (cloning required)
- Tet-ON worked on/off upon Dox addition (no dose dependence)
Stop codon suppression
- Protein X expression is blocked by adding a stop codon in the coding sequence
- An engineered suppressor tRNA restores lower-level expression
- GOI (Protein X) expression is also restored by geneticin in a dose-dependent manner
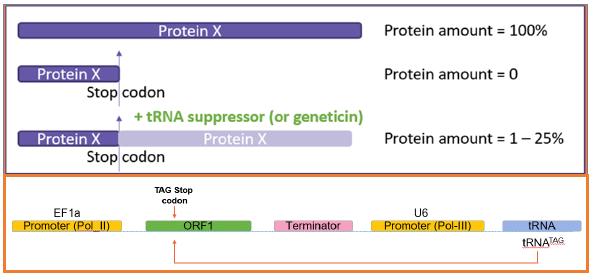
Image Credit: Image courtesy of Marina Fedorova and Ian Hiles in partnership with ELRIG (UK) Ltd.
Conditions:
- CHO GSKO cells
- Transfection: expression cassette vectors co-transfected with mRNA transposase
- Protein X expression: flow cytometry
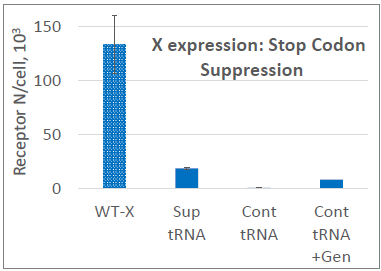
Image Credit: Image courtesy of Marina Fedorova and Ian Hiles in partnership with ELRIG (UK) Ltd.
Control tRNA: no expression
Suppressor tRNA: low Protein X expression
Control tRNA + Geneticin: low Protein X expression
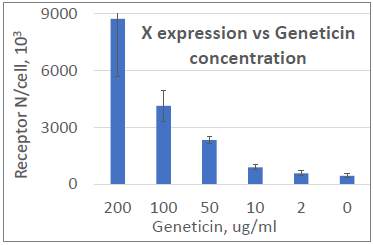
Image Credit: Image courtesy of Marina Fedorova and Ian Hiles in partnership with ELRIG (UK) Ltd.
Geneticin restores Protein X expression in a dose-dependent manner
Learnings
- Protein X below 20 K receptors
- Geneticin induces titratable stop codon readthrough
Panelof weak promoters and DNA modifications
11 Protein X and 7 Protein Y constructs (co-transfected with transposase mRNA)
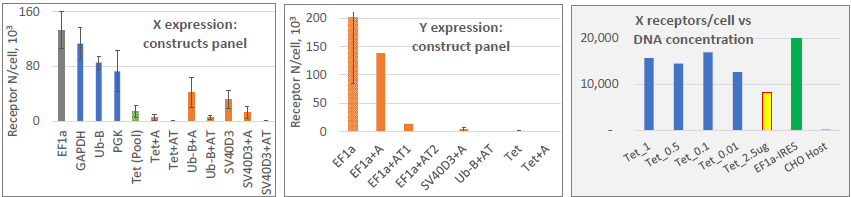
Image Credit: Image courtesy of Marina Fedorova and Ian Hiles in partnership with ELRIG (UK) Ltd.
- 13 out of 18 constructs had non-expressing population peaks
- DNA concentration didn’t change the expression (range: 0.01-9 ug/transfection)
- Required Protein X expression (~15 K) was achieved under Tet promoter
- But: ~6-11 % non-expressing population peak, growing – required cloning
Conditions:
- CHO GSKO cells
- DNA + transposase mRNA
- Protein X/Y expression: flow cytometry
CellCelector™ Flex Cloning: Tet-Protein X
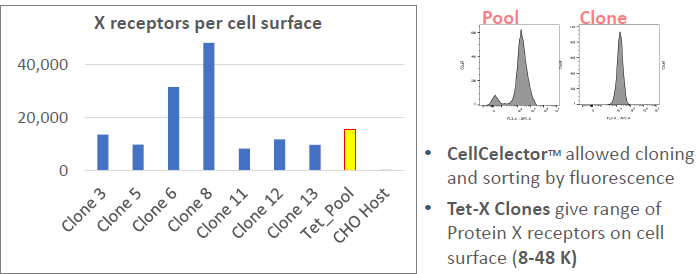
Image Credit: Image courtesy of Marina Fedorova and Ian Hiles in partnership with ELRIG (UK) Ltd.
Learnings
- Range of Protein X expressions (1-150 K receptors)
- Further single cell cloning is required (performed with CellCelector™ Flex)
IRES element prior gene of interest
- IRES prior gene of interest reduces expression
- ORF 1: Glutamine Synthetase selection; ORF 2: Protein X or Y

Image Credit: Image courtesy of Marina Fedorova and Ian Hiles in partnership with ELRIG (UK) Ltd.
Conditions:
- CHO GSKO cells
- DNA + tranposase mRNA
- Protein X/Y expression: flowcytometry
“Classic” IRES element
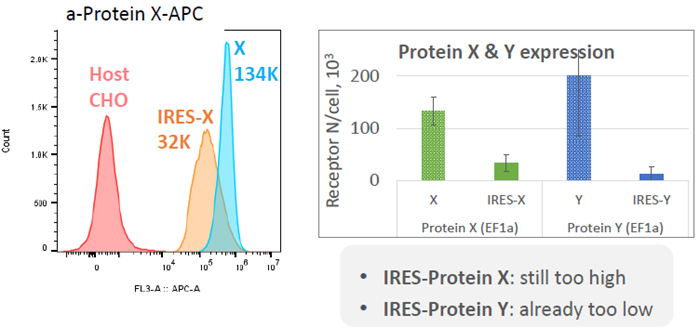
Image Credit: Image courtesy of Marina Fedorova and Ian Hiles in partnership with ELRIG (UK) Ltd.
Modified IRES element
Varying sequence around the 3 x ATG codons at the end of the IRES impacts expression.
Aim: Slightly increase IRES-Protein Y expression
Constructs:
- Change the 7 x A sequence present in the bi-furcation loop to 6 x A - should increase expression ~10 fold)
- Move the ATG start codon for Protein Y to the optimal position in the IRES sequence (should increase expression)
- Combine both the above in one plasmid (should give cumulative increase in expression)
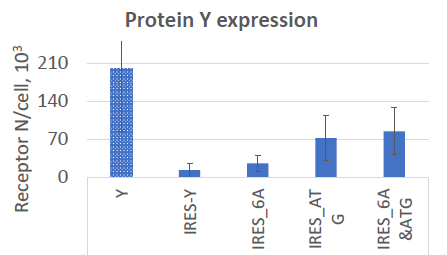
Image Credit: Image courtesy of Marina Fedorova and Ian Hiles in partnership with ELRIG (UK) Ltd.
Learnings
- Classic IRES: Protein X levels to 30 K, Protein Y levels to 13 K
- Modified IRES increased Protein Y levels to ~25 K
- Different “strength” IRES can predict target expression
Summary and conclusions
- Four methods to tune protein expression were developed and optimized
- Cells for the program were generated using 2 of these methods
- Promoter panel (Protein X)
- Modified IRES elements (Protein Y)
- Methods can be chosen depending on the program requirements, timelines and resources
Source: ELRIG (UK) Ltd.
| |
|
Receptors/cell |
Ratio |
| |
Cells |
Protein X |
Protein Y |
X : Y |
| Original |
Protein X |
100 K-1300 MIL |
|
|
| Protein Y |
|
100 K-300 K |
|
| Protein X & Y |
- |
- |
|
| Tuned down |
Protein X |
9.4 K |
|
|
| Protein Y |
|
21.1K |
|
| Protein X & Y |
10 K |
23.8K |
1:2.4 |
Source: ELRIG (UK) Ltd.
| |
Summary of method |
Details/limitations |
| Tet-ONE |
Dox-inducible ON/OFF low expression |
No dose-dependent response to Dox, requires cloning |
| Stop Codon |
Low levels and precise titratable .expression |
In-house cassettes: vectors can be slotted in |
| Construct Panel |
Wide range of expressions |
Customize each time at CRO – takes time, requires .cloning |
| IRES |
Precise tuning up and down |
Different strength IRES can be developed further |
Acknowledgments
Many thanks to Ian Hiles and Stefano Tonin for molecular biology design and construct generation. Thank you to Paul White and Program team for their suggestions; to Danny Acquisto, Lindsey Pearson and Sartorius team for help with CellCelector™ Flex; to Lorena Guglielmo for support in cell scale-up; CSF team (Sam Payne and Glenn Lewis) for keeping all these cell lines alive and happy and to Cellular Technology team for their support.
References
- Wang, J., et al. (2022). AAV-delivered suppressor tRNA overcomes a nonsense mutation in mice. Nature, [online] 604(7905), pp.343–348. https://doi.org/10.1038/s41586-022-04533-3.
- Koh, E.Y.C., Ho, S.C.L., Mariati, Song, Z., Bi, X., Bardor, M. and Yang, Y. (2013). An Internal Ribosome Entry Site (IRES) Mutant Library for Tuning Expression Level of Multiple Genes in Mammalian Cells. PLoS ONE, [online] 8(12), p.e82100. https://doi.org/10.1371/journal.pone.0082100.
About GSK
GSK are a focused biopharma company. They prevent and treat disease with vaccines, specialty and general medicines. They focus on the science of the immune system and advanced technologies, investing in four core therapeutic areas: infectious diseases, HIV, respiratory/immunology, and oncology.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 18, 2024