This article is based on a poster originally authored by Sophie Snow, Kira Freeman, Zaynab Isseljee, Alice Brankin and Christine Sanfelice, which was presented at ELRIG Drug Discovery 2024 in affiliation with LifeArc and University of Bath.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Bridging the gap between academic research and unmet patient need
LifeArc is a non-profit medical research charity that supports early-stage research through partnerships, translational expertise, and investment.
Translational challenges are under-served areas within the clinic that LifeArc is prioritizing:
- Motor Neuron Disease
- Chronic Respiratory Infections
- Global Health
- Rare Diseases
- Childhood Cancer
LifeArc has several in-house capabilities set up to support translational science projects:
- Antibody discovery, development and humanization
- Small molecule development
- Microbiology
- Induced pluripotent stem cells
- Cheminformatics and bioinformatics
Introduction
Early identification of diverse candidate molecules is key in the antibody drug discovery process.
Antibodies that bind to similar target epitopes often share similar functions. Therefore, identifying distinct epitope bins with functional activity can potentially assist in generating a reduced panel of epitope-diverse candidates to select from, with different modes of action on the antigen.
As highly specific probes capable of correlating structural determinants with functional properties, monoclonal antibodies (mAbs) also serve as useful tools in validating drug targets.1 For example, identifying antibody pairs that can simultaneously bind the antigen enables their use as reagents for downstream applications that require this sandwiching interaction.2
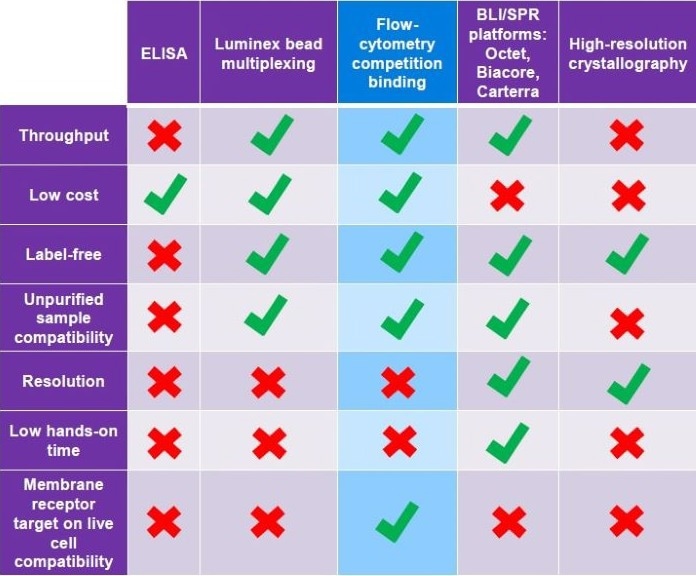
Table 1. A comparison of epitope binning methods used at various stages of therapeutic candidate selection. Each technique has unique advantages and disadvantages depending on the stage of selection. E.g. low cost but low-resolution ELISA is suitable during early screening, whilst high-resolution but expensive crystallography is better utilised when homing in on a final lead candidate. Source: ELRIG (UK) Ltd.
Assay principle
Flow cytometry competition binding is a relatively novel characterization assay designed to enable epitope binning using the competitive binding profiles of monoclonal antibodies to protein targets.
In principle, monoclonal antibodies with competitive binding profiles against a defined set of reference antibodies will likely bind to the same site on a target antigen, indicating similar epitopes.
By saturating the target antigen with pooled reference antibodies, the epitope sites specific to those antibodies are effectively blocked, preventing further binding by additional antibodies that share the same binding sites and sit within the same epitope space. Antibodies that bind to alternative epitopes demonstrate additional binding and are classified as distinct from the reference antibodies and sorted into a different epitope bin.
This strategy means a large antibody panel can be accurately and quickly binned without requiring exhaustive antibody pairing interactions by determining the degree of correlation between binding profiles.
Experimental objectives
- To adapt this assay for screening a pilot panel of 48 antibody candidates against a diverse set of 15 reference antibodies, selected based on flow binding profiles and performance in a reporter cell assay. Modifications to the method described in 2018 by Chan et al.2 include alternative barcoding, a viability dye, and a newly developed script for automating the analysis of the raw data and generating figures.
- The results will be analyzed to yield epitope bins or clusters for the full antibody panel, creating a conceptual map of the epitope space for each antibody.
Method

Figure 1. An overview of the protocol for generating competitive binding profiles by flow cytometry for epitope binning. Cellular barcoding increases the number of reference antibody sets that can be run simultaneously. Individual barcoded cell populations, precoated with saturating concentrations of reference antibody, are then multiplexed with other barcoded reference antibody cell populations. Sample preparation prior to acquisition, including all incubation steps and washes, takes approximately 4 hours 30 minutes. Image Credit: Image courtesy of Sophie Snow et al., in partnership with ELRIG (UK) Ltd.
Analysis
Median Fluorescence Intensity (MFI) was calculated for the live population of each test antibody well against each reference set, performed within Forecyt®. This data was imported into an automated pipeline for analysis, which subtracted the MFI signal generated by the reference antibodies from the total MFI signal of each sample well. This enables quantifying the increase in binding signal and subsequent generation of competition binding plots (Figure 2).
The pipeline then compared the increase in signal for each test antibody against each reference antibody and calculated Pearson’s correlation coefficient for all pairings. A matrix plot of the correlation coefficients was produced and color-coded to create a heat map. The heat map was then sorted to assist in identifying clustering patterns (Figure 3), demonstrating the degree of similarity between all antibody pairings.
From this clustering analysis, the pipeline generated a dendrogram (Figure 4) showing the relationships between test antibodies within the epitope space. The distance between antibodies represents a relative measure of the difference in epitope binding site.
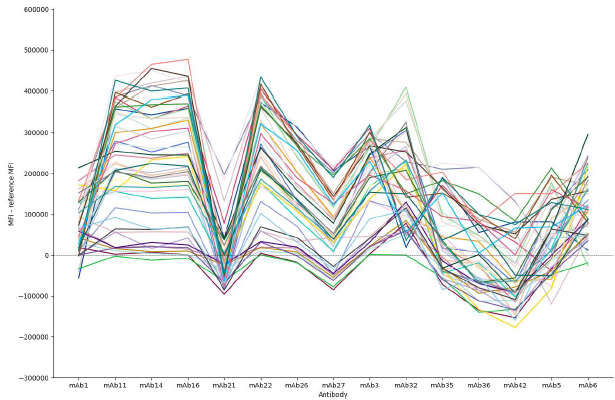
Figure 2. Binding profiles of 48 candidate antibodies against 15 reference antibodies. Binding profiles are generated from the additional fluorescence signal generated by the test antibodies for all combinations of reference antibody and test antibody. Similar profiles indicate shared epitope binding sites on the target antigen and can be classified together. By comparing binding profiles rather than individual data points, differences due to antibody concentration or affinity are not mistaken for a different epitope bin, as the binding profile would remain similar and just be variable in binding magnitude. Image Credit: Image courtesy of Sophie Snow et al., in partnership with ELRIG (UK) Ltd.
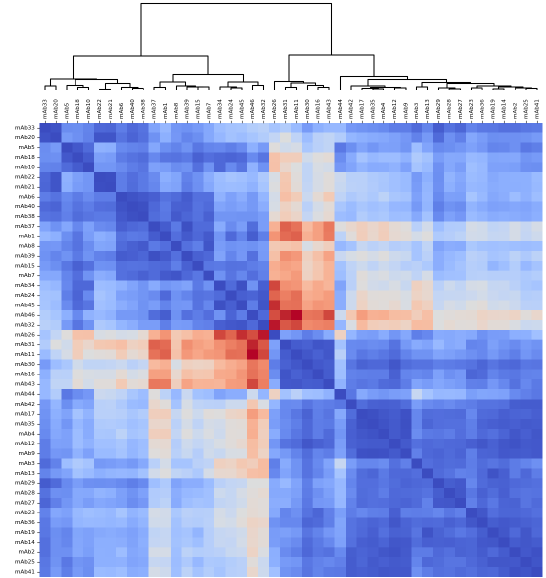
Figure 3. A heat map of the Pearson’s correlation coefficient values for all antibodies, sorted by similarity to identify clustering patterns. Dark blue coloration indicates a high degree of correlation in binding between antibody pairings (>90 %), suggesting the antibodies compete for the same binding sites on the antigen. Dark red coloration indicates a low degree of correlation in binding between antibody pairings (<10 %), suggesting the antibodies do not compete and therefore bind to alternative sites. Antibodies that share similar correlation values across the panel will likely be sorted into the same epitope bin. Image Credit: Image courtesy of Sophie Snow et al., in partnership with ELRIG (UK) Ltd.
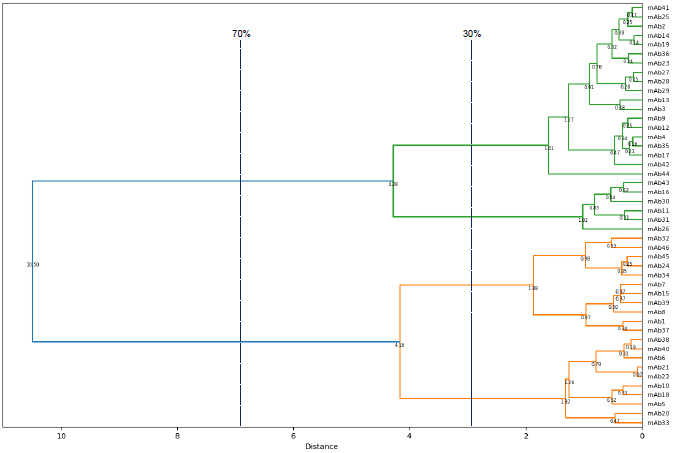
Figure 4. A dendrogram displaying the relationships of similarity in epitope binding sites between antibodies from the panel. Two main branches are observed at 70 % of the greatest distance within the epitope space, i.e. the distance between the two most distantly related antibodies within the panel. These sub-divide into four branches at 30 % of the total distance, which enables antibodies from the panel to be classified into one of four epitope bins. The assignment of this panel of antibodies within these bins was validated by Surface Plasmon Resonance (SPR) methods including Carterra® LSA™ and Biacore™ platforms (data not shown). Image Credit: Image courtesy of Sophie Snow et al., in partnership with ELRIG (UK) Ltd.
Discussion and conclusions
This pilot study has found that epitope binning by way of comparing competition binding profiles can accurately classify a panel of monoclonal antibody candidates relative to each other, generating results supported by current gold standard methods. Therefore, this flow cytometry-based method provides significant advantages in early-stage therapeutic molecule screening, enabling high throughput panel characterization and rational lead candidate selection without extensive cost or sample compatibility limitations.
The granularity of epitope binning can be adjusted based on the degree of resolution required. By optimizing this protocol, we have found that a large reference antibody set improves the resolution of the epitope bins. However, the number of references can be reduced without detriment to accurate classification. This is on the provision that the reference set provides sufficient epitope diversity, which can be determined with a scouting experiment. The cut-off threshold for assigning bins can be chosen based on the level of differentiation desired or as a percentage of the most significant distance within the epitope space.
A potential limitation of this assay is the lack of resolution due to steric hindrance between antibodies specific to neighboring epitopes. Competition for the same binding site cannot be differentiated from obstruction of adjacent sites, brought about by the large size of these molecules. The possibility of this effect occurring and the way to negate it depends upon the precise combinations of reference antibodies within each set and may, therefore, necessitate testing candidates against additional combinations of reference antibodies to ensure sufficient diversity between the sets. Using Fab regions instead of full-length antibodies may also serve as a solution.
Further optimization work will be performed to explore the impact of antibody dissociation rates on binning results and refine the cell-antibody preparation steps to reduce the overall length of the protocol.
References
- Jia, X.-C., et al. (2004). A novel method of Multiplexed Competitive Antibody Binning for the characterization of monoclonal antibodies. Journal of immunological methods, 288(1-2), pp.91–98. https://doi.org/10.1016/j.jim.2004.02.010.
- Chan, B.M., et al. (2018). Flow Cytometry-Based Epitope Binning Using Competitive Binding Profiles for the Characterization of Monoclonal Antibodies against Cellular and Soluble Protein Targets. SLAS DISCOVERY: Advancing the Science of Drug Discovery, 23(7), pp.613–623. https://doi.org/10.1177/2472555218774334.
About Life Arc
Life Arc fund, collaborate and provide scientific expertise across some of the most under-served conditions: motor neuron disease, chronic respiratory infection, global health, rare disease, and childhood cancer.
They look to invest in projects where they can use their experience in translational science to remove barriers for early scientific discoveries.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 18, 2024