From NucleraReviewed by Olivia Frost
This article is based on a poster originally authored by Chava Angell, Eric Swanson, Andrea Zanzotto, and Yvonne Tan, which was presented at ELRIG Drug Discovery 2024 in affiliation with Nuclera Ltd.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

E3 ligases transfer ubiquitin to target proteins for degradation, requiring precise recognition of both ubiquitin and their substrate. This specificity makes E3 ligases valuable drug targets for manipulating degradation pathways to restore cellular function or remove harmful proteins. The ability to rapidly generate soluble, active recombinant E3 ligases is crucial for drug discovery targeting the ubiquitin pathway.
This study demonstrated that eProtein Discovery enables protein scientists to rapidly screen and identify the optimal constructs and conditions to generate soluble recombinant E3 ligases Cereblon, VHL, and Parkin.
Method
The eProtein DiscoveryTM system enables multiplexed screening to evaluate the following constructs and conditions for expression simultaneously:
- Length variants: Multiple protein length variants were designed guided by AlphaFold2 3D predicted model
- Solubility tags: DNA constructs were expanded to include different solubility tags (ZZ, TRX, SUMO) and without
- Detagging: In situ removal of solubility tags and evaluate the impact on protein yield
- Expression conditions: Rapid cell-free expression screen with different additives
eProtein Discovery workflow
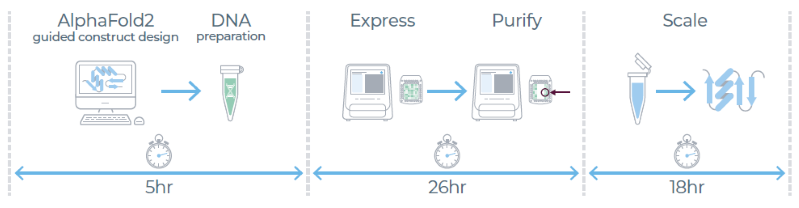
Image Credit: Chava Angell et al., in partnership with Nuclera Ltd.
Result 1
Protein target: Cereblon
Constructs designed based on the AF2 model:
- Full-Length Cereblon
- Cereblon Δ1-42:Cereblon with residues 1-42 removed
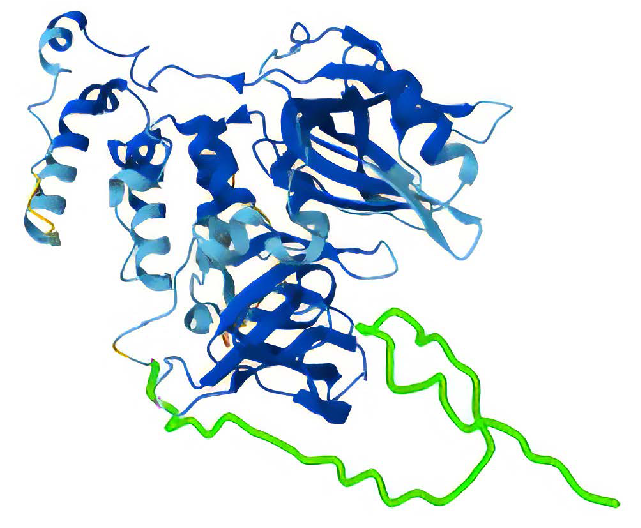
Structure of full-length Cereblon. In green, residues 1-42 was removed in construct Cereblon Δ1-42. Image Credit: Chava Angell et al., in partnership with Nuclera Ltd.
Purified yield:
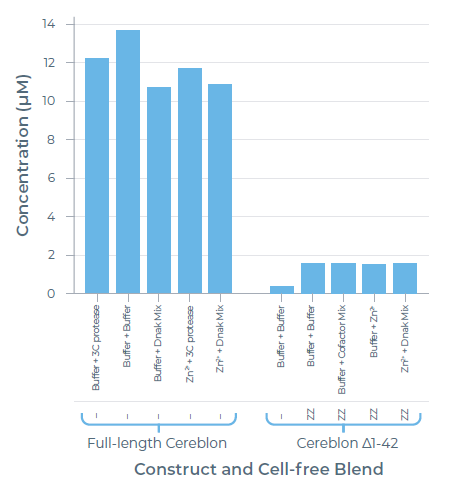
Image Credit: Chava Angell et al., in partnership with Nuclera Ltd.
Result 2
Protein target: von Hippel-Lindau (VHL)
Constructs:
- Full-Length VHL
- VHL Δ1-57: VHL with residues 1-57 removed
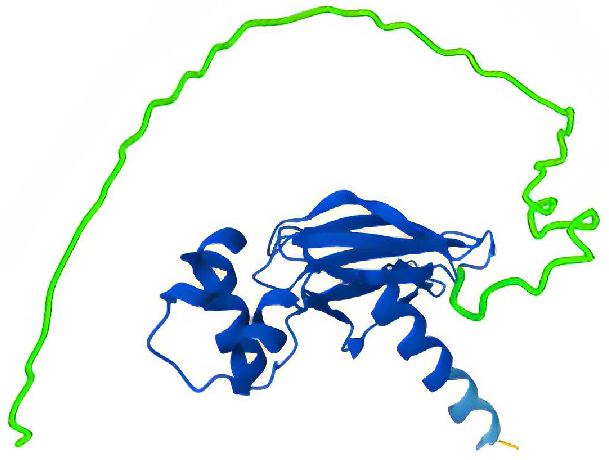
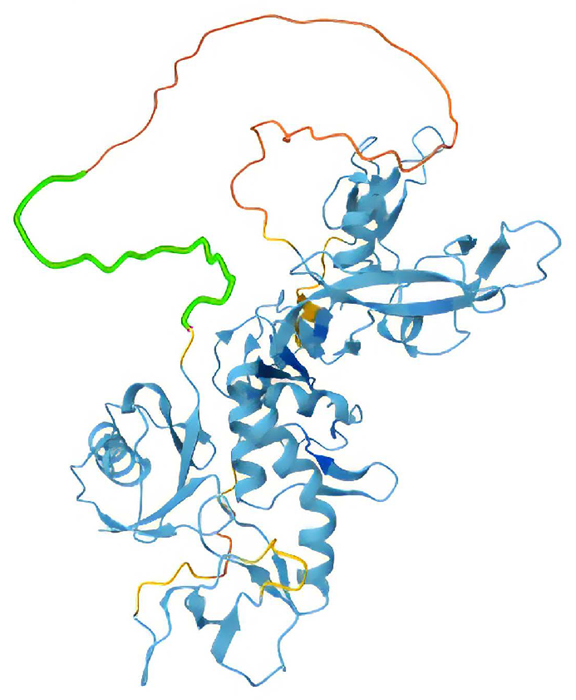
Structure of full-length VHL. In green, residues 1-57 was removed in construct VHL Δ1-57. Image Credit: Chava Angell et al., in partnership with Nuclera Ltd.
Purified yield:
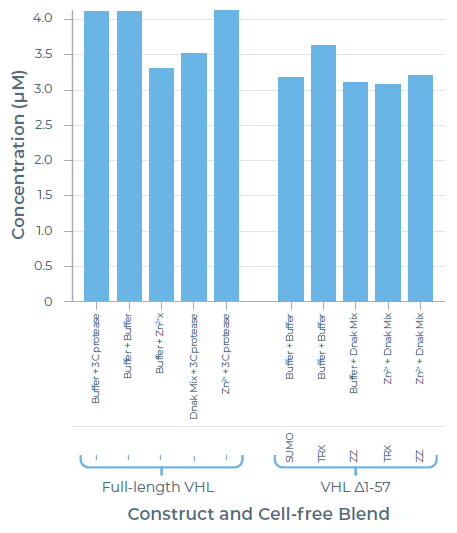
Image Credit: Chava Angell et al., in partnership with Nuclera Ltd.
Result 3
Protein target: Parkin
Constructs:
- Full-length Parkin
- Parkin Δ76-98: Parkin with partial loop removed
- Parkin Δ84-143: Parkin with complete loop removed
Structure of full-length Parkin, in green, construct Parkin Δ76-98 with midloop residues removed. Image Credit: Chava Angell et al., in partnership with Nuclera Ltd.
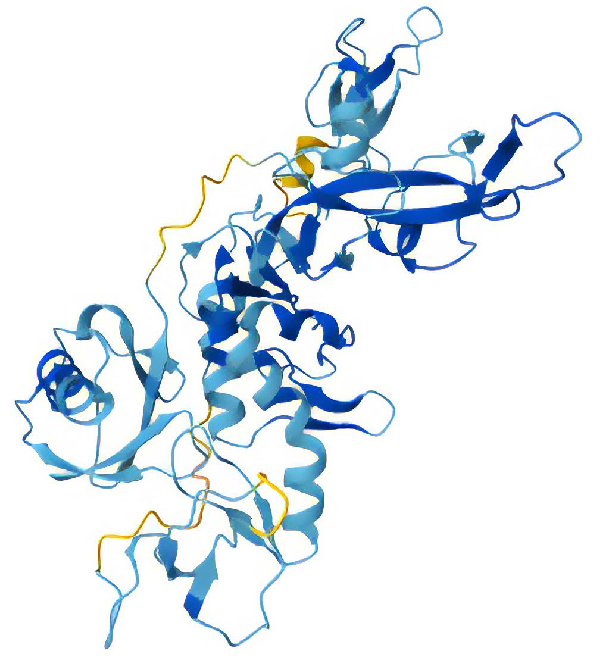
Structure of Parkin Δ84-143 with the entire loop removed. Image Credit: Chava Angell et al., in partnership with Nuclera Ltd.
Purified yield:
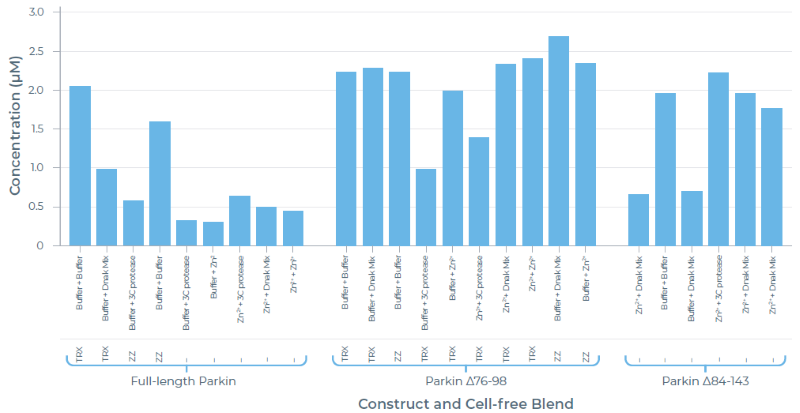
Image Credit: Chava Angell et al., in partnership with Nuclera Ltd.
Conclusion 1
- Full-length Cereblon without solubility tags gave the highest purified yield.
Conclusion 2
- Removing the disordered N-terminal residues did not significantly increase protein expression and purification.
- The solubility tag can be removed with 3C protease if tags are not tolerated in downstream assays.
Conclusion 3
- Parkin Δ76-98 with the small disordered region removed gave the highest purified yield.
- Interestingly, removing the entire loop did not further increase protein yield.
Summary
The eProtein DiscoveryTM system has proven to speed up recombinant E3 ubiquitin ligase production by rapidly screening multiple constructs and conditions in just one day, delivering actionable insights for scaling up and advancing targeted protein degradation drug discovery.
About Nuclera
Nuclera is the pioneer in bringing rapid protein prototyping to the benchtop. We make proteins accessible through eProtein Discovery™.
Through our technology, we accelerate breakthrough improvements in human health and empower life science researchers with easy access to target proteins.
Founded in 2013 by PhD students at the University of Cambridge, the company now has a presence in Cambridge, UK and Boston, MA employing over 100 employees.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 18, 2024