This article is based on a poster originally authored by Barbie Wang, Maria Giebler, Adrian Freeman, Karen Hogg, Adam Corrigan and Hitesh Sanganee.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Abstract
Imaging flow cytometry (IFC) enables the identification of cells with the throughput of a traditional flow cytometer, with added imaging features enabling single cell image acquisitions and cell sorting based on phenotypes. We collaborated with the imaging facility at the University of York to access BD FACSDiscover S8 imaging flow cytometry system to explore the potential of Phenospace as an AI tool to determine patient cellular phenotypes.
The generation of cellular images and transcriptomic data from respective sorted cell populations allows us to train a machine learning platform to exclusively identify transcriptomic changes within cells based solely on the analysis of images. This work establishes crucial groundwork, yielding multiple proof of concept data for advancing this methodology toward clinical applications.
Introduction
- IFC allows for real-time visualization of cell-cell interaction, co-localization, and cell phenotypic changes. We generate images from drug-treated primary immune cells and underpin the transcriptomic changes using NGS of sorted populations.
- To mimic disease cell states, we used IFNa as a stimulant to induce a phenotype known from systemic Lupus Erythematous (SLE), while Saphnelo (INFAR-inhibitor) was used to restore cells to a healthy phenotype. Leveraging our phenotypic data, we fed it into an in-house machine learning image analysis tool, Phenospace, to generate imaging profiles that could be linked to respective transcriptomic profiles.
Methods
Image Credit: Image courtesy of Barbie Wang et al., in partnership with ELRIG (UK) Ltd.
Results
A: Gating strategies allow for identification of distinct cell types based on flow parameters
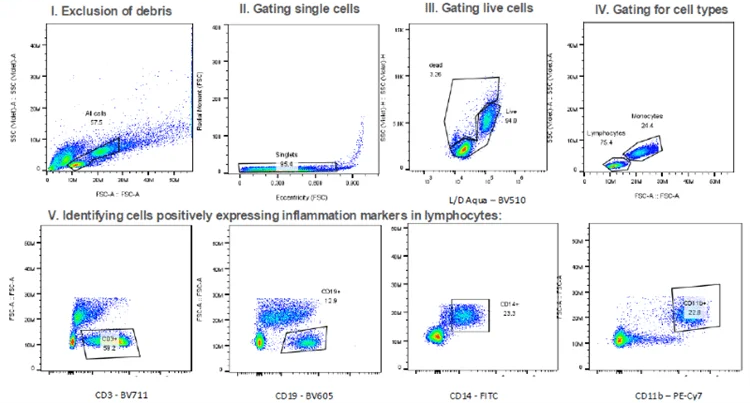
Image Credit: Image courtesy of Barbie Wang et al., in partnership with ELRIG (UK) Ltd.
In (V), the relevant surface expression markers were analysed to determine the effect of IFNa and Anifrolumabs on different cell types. Positive CD3 expression allows for selection of T cells, CD19 for B-cells while CD14 and CD11b are used as inflammatory markers.
B. Analysis of expression and DEGs
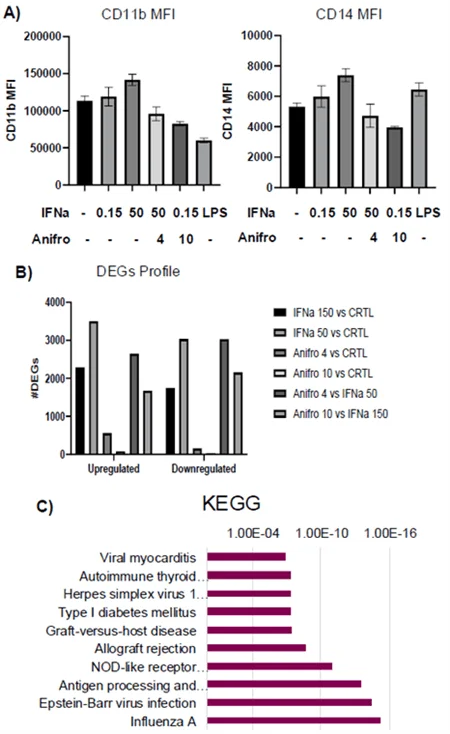
A) Bar graphs created using GraphPad, showcasing expression of CD11b and CD14 in PBMCs upon stimulation with IFNa and Anifrolumab. B) Comparison between the number of upregulated vs downregulated genes under different treatment conditions. DEGs were generated from sorted live cells as seen in gating strategies (III). Samples treated with Anifrolumab can be seen to have minimal DEGs, evidencing success in returning cells to healthy state. C) KEGG showing the least to most significant pathways extracted from top 500 DEGs from 150 ng IFNa treated samples compared to control samples. Image Credit: Image courtesy of Barbie Wang et al., in partnership with ELRIG (UK) Ltd.
Future opportunities
Phenospace, an in-house ML image analyzing tool differentiates between the different cellular phenotypes, allowing clustering of the same cell types based on images. The next step is to evaluate whether it can distinguish between different cell states. QC of Phenospace predictions can be done through the generation of confusion matrices.
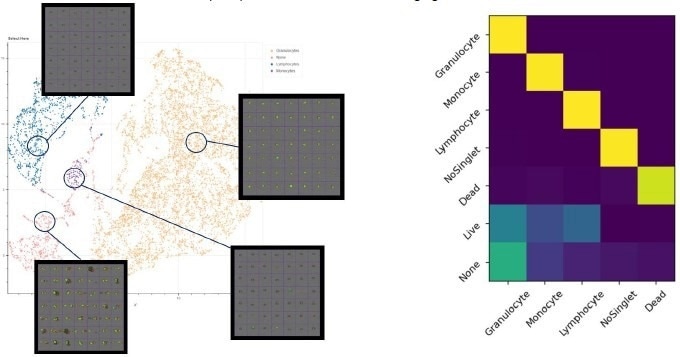
Image Credit: Image courtesy of Barbie Wang et al., in partnership with ELRIG (UK) Ltd.
Conclusion and next steps
- Generated multiple proof-of-concept data, including images and transcriptomic data.
- Train Phenospace using this batch of data to see if it can distinguish between stimulated and unstimulated cell types based on images.
- Utilize artificial intelligence (AI) to aid in the discovery of new compound indications and to distinguish responding from nonresponding patient samples.
Acknowledgments
Maria Giebler, Adrian Freeman, Agnieszka Jozwik, Karen Hogg, Adam Corrigan, Sukhveer Mann, Wider EIU and FGx team.
About AstraZeneca
AstraZeneca is a global, science-led patient-focused pharmaceutical company that focuses on transforming the future of healthcare by unlocking the power of what science can do for people, society and the planet.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 18, 2024