This article is based on a poster originally authored by Ebru Aydin Kurtulmus, Dagmar Hildebrand, Fiordiligie Casilag, Verena Banschbach, Jessica Heckmann, Sarah Schott, Marc Botcherby and Volker Stadler, which was presented at ELRIG Drug Discovery 2024 in affiliation with PEPperPRINT GmbH.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Aim
Given the growing numbers of therapeutic protein products (TPPs) being developed, the generation of tools to identify immune responses mounted against them has become significantly important. At PEPperPRINT, we developed a novel pipeline to enable:
- Profiling potential T-cell immunogenicity through chip-based MHC-associated peptide proteomics (MAPPs) assay
- Discrimination of regulatory T-cell epitopes from effectors and assessment of immunogenicity risk based on epitope content
- Identification of immunogenic epitopes recognized by anti-drug antibodies (ADA)
- Monitoring of pre-existing and treatment-emergent antibody responses for assessment of treatment risks and immunogenic adverse effects
Study background
In this study, we adapted our peptide microarray platform technology to assess the immunogenic potential of two extensively studied PD-L1 inhibitors, which exhibit markedly different reported immunogenicities. We aim to validate existing clinical data.
Therapeutic protein products have revolutionized the treatment of several diseases, including cancer, chronic autoimmune, and inflammatory diseases. However, many of these therapeutics are reported to be immunogenic at various levels.1 This immunogenicity is associated with ADA generation, which can neutralize drug efficacy by altering its pharmacokinetic and pharmacodynamic properties. Above all, ADAs may cause several immune adverse effects in patients.
Discovering biomarkers that identify patients at risk is essential to design safer drugs with lower rejection rates. The immunogenicity of TPPs can manifest in the form of different immune responses depending on the patient’s T-cell and B-cell repertoires, cytokine milieu, and prior exposure of the immune system to proteins of similar structure. Identification of ADA epitopes in polyclonal Ab mixtures like sera is usually challenging due to this high diversity. Here, we show that high-density peptide microarrays are powerful tools to simultaneously screen thousands of peptides against serum antibodies in a high-throughput context.2
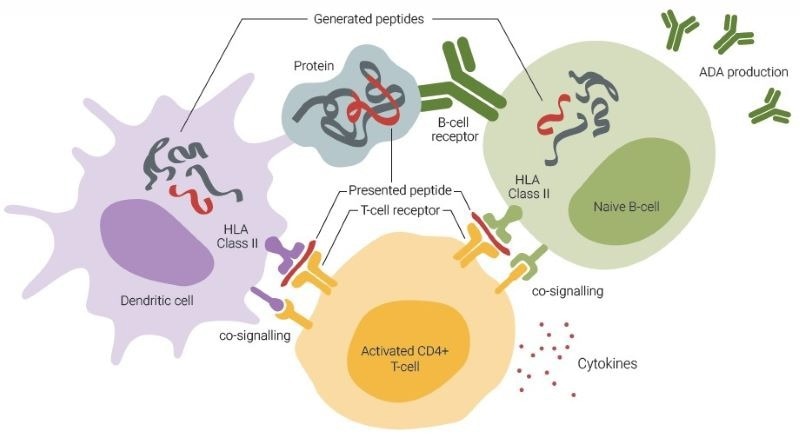
Fig. 1. ADA formation involves an interplay among dendritic cells (DCs), T-cells, and B-cells. DCs primarily capture antigens through macropinocytosis and receptor-mediated uptake, enabling them to sample extracellular proteins. Conversely, B-cells predominantly internalize antigens by recognizing their specific structures via B-cell receptors. DC-activated T-cells can subsequently activate B-cells that are presenting identical sequences. Image Credit: Image courtesy of Ebru Aydin Kurtulmus et al., in partnership with ELRIG (UK) Ltd.
Although ADAs have historically been a standard clinical measure of immunogenicity, the root cause is the T-cell dependent (Td) immune response generated through the presentation of epitopes by subsets of human leukocyte (HLA) class II molecules to CD4+ T-cells that drive B-cell maturation and ADA generation.
Current practice of immunogenicity screening starts with an assessment, followed by in vitro and ex vivo analyses such as HLA binding assays, T-cell assays, and MHC-associated peptide proteomics (MAPPs).
The MAPPs assay involves multiple steps involving human primary antigen-presenting cell culture, peptide isolation, and peptide identification via liquid chromatography-mass spectrometry (LC-MS)3. Clinical validation of the MAPPs assay remains challenging due to limitations in instrument sensitivity, labor-intensive processes, and the requirement for substantial amounts of patient material, in addition to dependence on predictions and complex computational methodologies. Here, we present a novel chip-based methodology that effectively surmounts these limitations, allowing for the discovery of epitope-specific T-cell responses in a remarkably short timeframe and eliminating the requirement for patient material.
Methodology and results
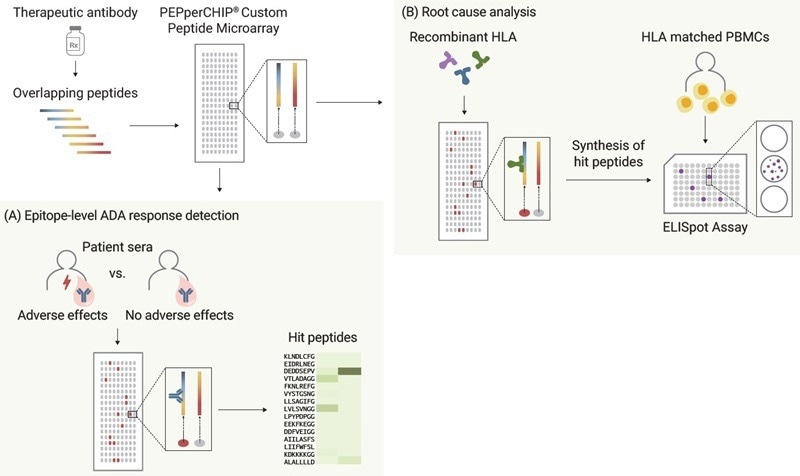
Fig.2. Amino acid sequences of two FDA-approved PD-L1 inhibitors, Durvalumab and Atezolizumab (see reference 4 for immunogenicity data) and generated a high-density peptide microarray by converting these amino acid sequences into 15 amino acid peptides with a maximum peptide-peptide overlap of 14 amino acids for high-resolution epitope data. (A) To detect the anti- drug antibody responses on the epitope level, serum samples from Druvalumab- or Atezolizumab-treated lung cancer patients are screened with PEPperCHIP® Peptide Microarrays. (B) For root cause analysis, same microarrays were analyzed using most common recombinant HLA-DR allotypes in global population, enabling the ranking of two PD-L1 inhibitors by immunogenic potential. Subsequently, identified peptide candidates are synthesized and applied in ELISpot/ FluoroSpot assays with HLA-matched PBMCs. Image Credit: Image courtesy of Ebru Aydin Kurtulmus et al., in partnership with ELRIG (UK) Ltd.
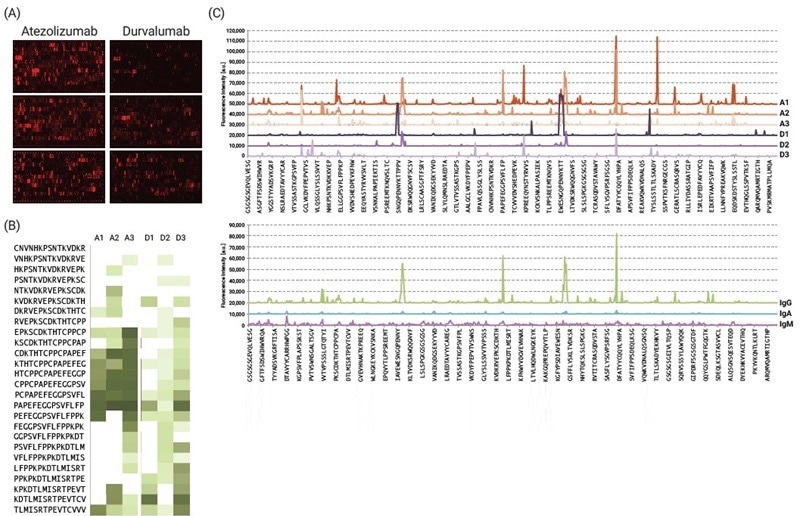
Fig. 3. IgG, IgM and IgA responses in Atezolizumab- or Durvalumab-treated lung cancer patients. Microrarrays were incubated with respective sera (1:100) overnight at 4 °C. (A) Sera from 3 representative cases of each treatment group. Detection of antibody binding was done using anti-human IgG DL680 (B) Identified hit peptides are presented as a heat map based on their reactivity, in addition to (C) intensity plots for immunoglobulin isotype/subtype analysis. Image Credit: Image courtesy of Ebru Aydin Kurtulmus et al., in partnership with ELRIG (UK) Ltd.
Result: While our study is based on limited sample size, PEPperCHIP® Microarrays enabled swift characterization of epitope-specific anti-atezolizumab ADA responses. IgA and IgM-mediated immune responses were scarce, whereas IgG was found to be the most relevant isotype in ADA analysis. Screening a larger cohort with synchronized sample collection is imperative for comprehensive evaluation.
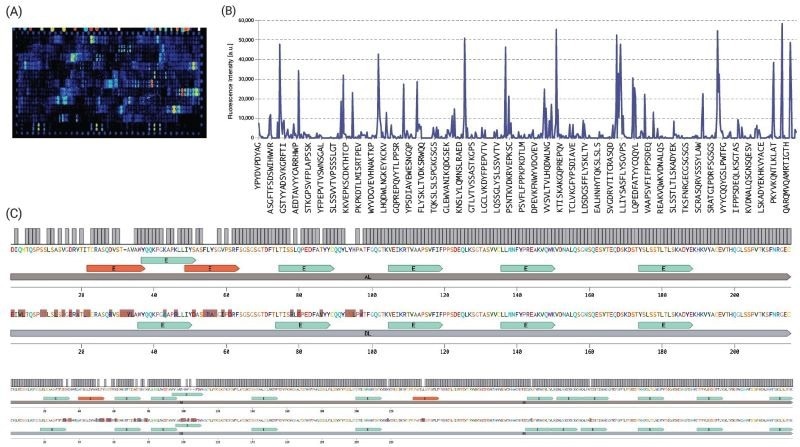
Fig. 4. For root cause/IRA analysis, the same microarrays were screened with recombinant HLA class II constructs, followed by a validation assay. Due to space limitations, here we show the screening results only for (A) HLA-DRB1:0101, which is known to be associated with the risk of developing ADA, and specifically NAb, in patients treated with Atezolizumab5. (B) The intensity plot highlights the interactions of recombinant soluble HLA-DRB1:0101 protein assayed at a concentration of 30 μg/ml. (C) To be able to visualize the immunogenic regions more clearly, we aligned the heavy and light chains of these inhibitors and integrated the epitope data from our chip-based MAPPS assay (blue). We found out 4 additional T-cell epitope candidates (red) on Atezolizumab amino acid sequence. These epitope candidates are being further validated in T-cell suppression and activation assays with the corresponding synthetic peptides. Image Credit: Image courtesy of Ebru Aydin Kurtulmus et al., in partnership with ELRIG (UK) Ltd.
Result: Our ranking aligns with existing literature, which indicates a significantly higher incidence of ADA formation for Atezolizumab (39.1 %) compared to Durvalumab (2.9 %).4 The inclusion of treatment-naive patients and healthy donors in the analysis would be advantageous for distinguishing preexisting and treatment-emergent ADAs.
Summary and key findings
In this study, we show the accuracy of the PEPperPRINT immunogenicity risk assessment pipeline in ranking two well-known PD-L1 inhibitors based on their epitope content.
Our innovative chip-based MAPPs methodology offers rapid identification of immunogenic hotspots of TPPs and presents several advantages over in silico and other in vitro MAPPs assays:
- The peptide library representing the TPP undergoes screening with recombinant human HLAs, obviating the necessity for patient samples or scarce biological material.
- Discovery occurs within a cell-free system, mitigating potential interference from cells or other protein-based culture materials.
- Testing of identified epitopes in AIM and Treg assays facilitates the assessment of their contribution to potential immunogenicity, yielding a more precise ranking.
- Peptide-HLA binding affinity data enhances the accuracy of immunogenic peptide ranking and provides guidance for drug development, without reliance on predictions or complex computational methodologies.
In addition to the chip-based MAPPs assay, the data generated with our ADA epitope mapping assay could be instrumental in:
- Assessing the contribution of immunogenicity to clinically observed adverse effects, if present.
- Determining whether the immunogenic response impacts drug efficacy.
- Identifying a unique epitope for patients experiencing specific side effects could aid in identifying individuals at higher risk of developing these adverse events.
References
- Vaisman-Mentesh, A., Gutierrez-Gonzalez, M., DeKosky, B.J. and Wine, Y. (2020). The Molecular Mechanisms That Underlie the Immune Biology of Anti-drug Antibody Formation Following Treatment With Monoclonal Antibodies. Frontiers in Immunology, [online] 11, p.1951. https://doi.org/10.3389/fimmu.2020.01951.
- Kearns, J.D., et al. (2023). A root cause analysis to identify the mechanistic drivers of immunogenicity against the anti-VEGF biotherapeutic brolucizumab. Science Translational Medicine, 15(681). https://doi.org/10.1126/scitranslmed.abq5068.
- Karle, A.C. (2020). Applying MAPPs Assays to Assess Drug Immunogenicity. Frontiers in Immunology, 11. https://doi.org/10.3389/fimmu.2020.00698.
- Davda, J., et al. (2019). Immunogenicity of immunomodulatory, antibody-based, oncology therapeutics. Journal for ImmunoTherapy of Cancer, 7(1). https://doi.org/10.1186/s40425-019-0586-0.
- Hammer, C. et al., Cli. Tra. Sci., 15(6) pp. 1393-1399 (2022).
About Shelford Scientific
Shelford Scientific provide consulting services for epitope mapping and related technology as well as microarray statistical analysis.
Official Innopsys distributor in UK and Ireland. Supplying and supporting Innoscan microarray scanners for expression analysis, cytogenetics, protein arrays, RPPA, glycan arrays and glycomics studies. The range also offers solutions for high content screening and phenotypic screening in manual and autoloader batch mode using Mapix microarray software.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 18, 2024