This article is based on a poster originally authored by Harrison H, Glen A , Andrews B, Bayle E, Duncan C, Fyfe M , Guest P, Rausch O , Swain C, Thomas B, Zemla-Brown A, Baric R, Chen Y, Froggatt H, Heise M, Moorman N, Morales N, Scobey T, Sheahan T , Taft-Benz S, Williams A, McElwee M, O'Connor L, Patel AH, and Szemiel AM, which was presented at ELRIG Drug Discovery 2024 in affiliation with STORM Therapeutics Limited, University of North Carolina, MRC-University of Glasgow Centre for Virus Research (CVR).
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

The RNA genome of SARS-CoV-2 contains a 5′ cap that facilitates the essential functions of translating viral proteins, protecting them from exonucleases, and evading the host immune response. Viruses, including coronaviruses, produce their own capping enzymes, which include RNA methyltransferases.
In this study, the authors describe the development and characterization of inhibitors of a coronavirus methyltransferase, NSP14. These agents prevent viral replication and expose the virus to the host immune response, leading to potent anti-viral cellular activity across a range of viruses, including those with pandemic potential.
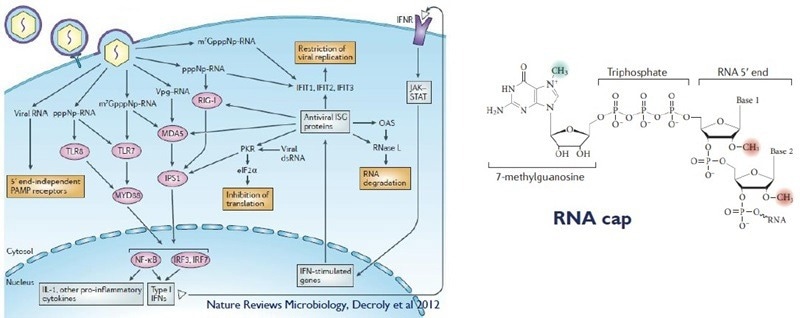
Image Credit: Harrison H et al., in partnership with ELRIG (UK) Ltd.
1) Identification & optimization of viral-specific inhibitors
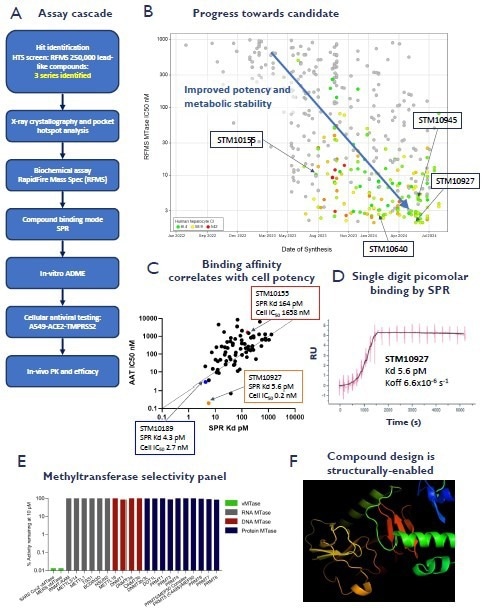
A. Schematic representation of the small molecule inhibitor profiling assay cascade. B. Graph illustrating the progress to make potent and metabolically stable analogs. C. Hit confirmation & routine design-make-test screening was performed using RapidFire Mass Spectrometry. Compounds too potent to accurately measure IC50s in the RFMS assay were tested in single-cycle kinetic surface plasmon resonance (SCK-SPR). SPR binding data has an excellent correlation with antiviral cell potency against SARS-CoV-2 in A549-ACE2-TMPRSS2 (AAT) cells. D. STM10927 has single-digit picomolar binding and a very slow off-rate by SCK-SPR. E. STORM methyltransferase inhibitors are virus-specific. The diagram shows the profiling of STM10155 in a human methyltransferase panel (Reaction Biology and STORM). Data are displayed as percent enzyme activity relative to DMSO-treated controls. F. Crystal structures of STORM compounds bound to the target are used for structure-based design. Image Credit: Harrison H et al., in partnership with ELRIG (UK) Ltd.
2) Compounds demonstrate good drug-like properties & potent cellular activity across the coronavirus family
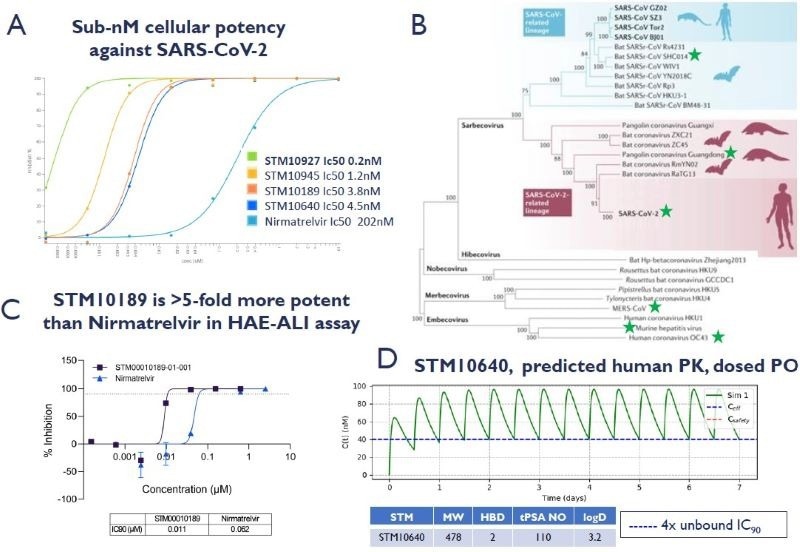
A. STORM compounds demonstrate significantly improved potency against SARS-CoV-2 in A549-ACE2- TMPRSS2 cells compared to Nirmatrelvir – the active component of Paxlovid (IF, SARS-CoV- 2/human/GBR/CVR-GLA-1/2020) B. Phylogenetic tree of Coronaviridae. STORM compounds demonstrate antiviral cellular potency for highlighted coronavirus subgenera: SARS-CoV-2, MERS, HuCoV-229E, HuCoV-OC43, PgCov/GD/2019, MHV and the emergent bat strain SHC14, with no cytotoxicity < 10 uM (A549-ACE2-TMPRSS2, Huh7, MRC-5, DBT, VeroE6 & Vero81 cells). Adapted from https://www.nature.com/articles/s41579-020-00459-7 C. STM10189 inhibition of SARS-CoV-2 human/USA/USA-WA1/2020 reporter infection in human airway epithelial cells grown at an air-liquid interface (HAE-ALI). D. STM10640 has a predicted oral dose of 8.5 mg/kg BID in human to cover 4x unbound IC90 at Cmin. Recent potent analogs have improved cell potency, human hepatocyte stability, and fraction absorbed in rodent in-vivo PK. Physicochemical properties are in drug-like property space. Image Credit: Harrison H et al., in partnership with ELRIG (UK) Ltd.
3) A novel platform for antiviral drug discovery with broad potential
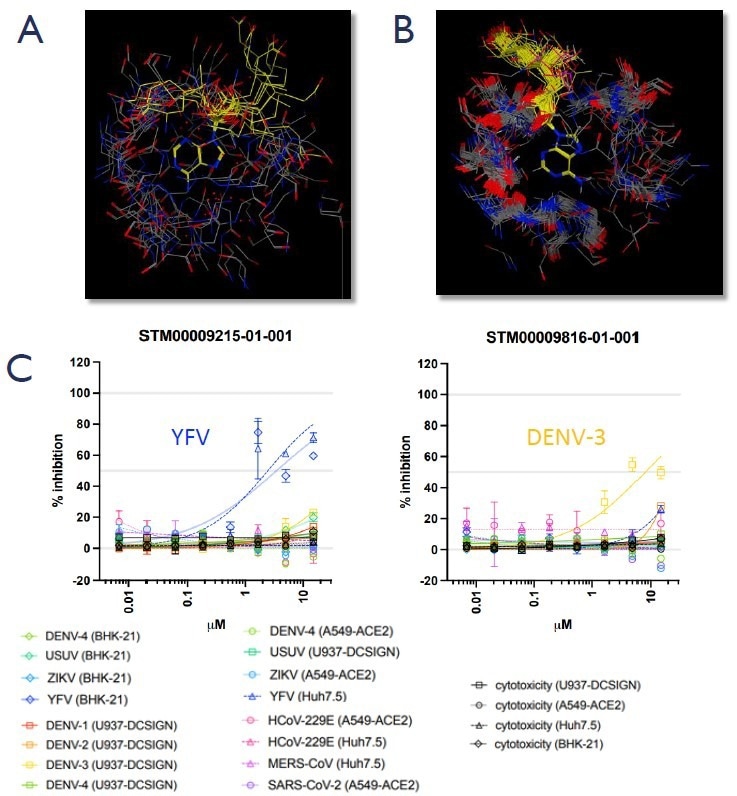
A. SAM/SAH, the methyltransferase cofactor/by-product, bound to a selection of non-viral human target proteins extracted from the PDB showing the diversity of conformations adopted in the proteins. B. SAM/SAH bound to all the viral methyltransferases extracted from the PDB showing the similarity of the cofactor conformation and virus binding pocket, demonstrating the potential for pan-viral activity. C. Preliminary data shows STM compounds specifically inhibiting viral replication of flaviviruses Yellow Fever and Dengue-3 in cell-based reporter assays, with no cytotoxicity up to 3 0 uM. Image Credit: Harrison H et al., in partnership with ELRIG (UK) Ltd.
Summary
- STORM has developed powerful NSP14 inhibitors with high selectivity for viral enzymes and cell potency that is 1000 times greater than the standard oral COVID-19 treatment.
- Structural and functional evidence suggests that STORM's viral methyltransferase inhibitors may also be effective against other viruses with pandemic potential and significant unmet medical needs.
- In-vitro and in-vivo tool compounds are now available to further explore the significant potential of viral methyltransferase inhibitors, leveraging their unique dual mechanism of action: anti-replicative activity and activation of the host's innate immune response against the virus.
About STORM Therapeutics LTD
STORM Therapeutics is a clinical-stage biotechnology company pioneering cellular reprogramming through RNA modifications to treat disease.
Its world leading understanding of RNA modifying enzymes (RME) has led to the discovery of breakthrough small molecule drugs that precisely reprogram cells through RNA biology for the treatment of cancer, inflammation, viruses and CNS diseases.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 18, 2024