This article and associated images are based on a poster originally authored by Tika R. Malla, Emma Mead and Emma J. Murphy and presented at ELRIG Drug Discovery 2024 in affiliation with University of Oxford.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Introduction
TREM2, a receptor predominantly expressed in microglia, regulates microglial proliferation, survival, migration, and phagocytosis, including clearance of amyloid plaques (one of the hallmarks of Alzheimer's disease).1 The activation of SHIP1, an inositol lipid phosphatase, is negatively linked to TREM2 signaling, lowering the neuroprotective effect of microglia.
A SHIP1 variant (rs39349669) confers risk for AD by increasing SHIP1 expression, inhibiting TREM2 signaling, and reducing the effector function of microglia.2,3 However, SHIP1 phosphatase has a very polar active site, making traditional biochemical screening approaches challenging or prone to high false positives.4 We therefore developed a cell-based degrader screen to target SHIP1 degradation.
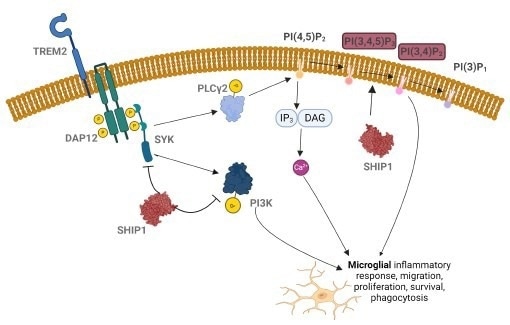
Figure 1. Inhibition of SHIP1 can upregulate phagocytic activity of microglia.2 Image Credit: Image courtesy of Tika R. Malla et al., in partnership with ELRIG (UK) Ltd.
Targeted protein degradation
Targeted Protein Degradation expands the druggable proteome beyond targets amenable to functional inhibition, e.g., transcription factors, scaffolding proteins, and seemingly difficult-to-drug enzymes like SHIP1. Coupling of the cellular ubiquitin proteasomal system (UPS) to a target protein of interest (POI) is achieved by Proteolysis Targeting Chimeras (PROTACs) or molecular glues that bring E3 ligases of UPS in proximity of the POI. Screening for serendipitous molecular glues is a promising way to discover novel anti-SIP1 mediated Alzheimer's therapeutics.
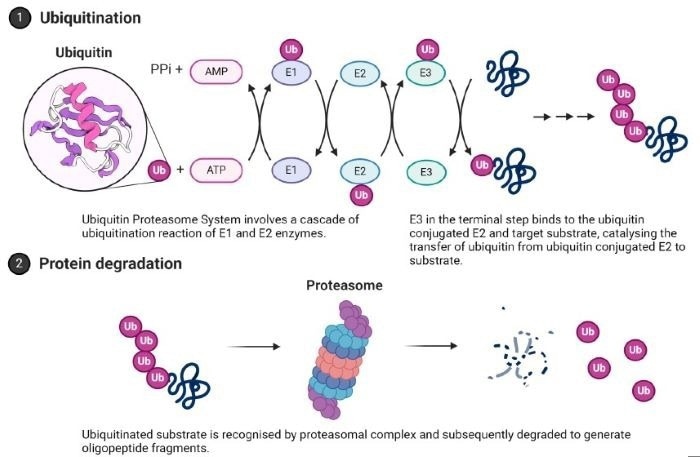
Figure 2. Ubiquitin Proteasome System (UPS). The terminal step that can be exploited in target protein degradation modality is highlighted. Image Credit: Image courtesy of Tika R. Malla et al., in partnership with ELRIG (UK) Ltd.
HT-bioluminescent assay
HiBiT LgBiT protein complementation can be used to monitor and conveniently quantify the abundance of endogenous protein. An 11 amino acid long HiBiT tag is added to the terminus of the POI by CRISPR editing of the target cell line. Live cell kinetic assays or endpoint luminescent assays can be performed to monitor protein abundance (Figure 3).
HiBiT-LgBIT complement assay
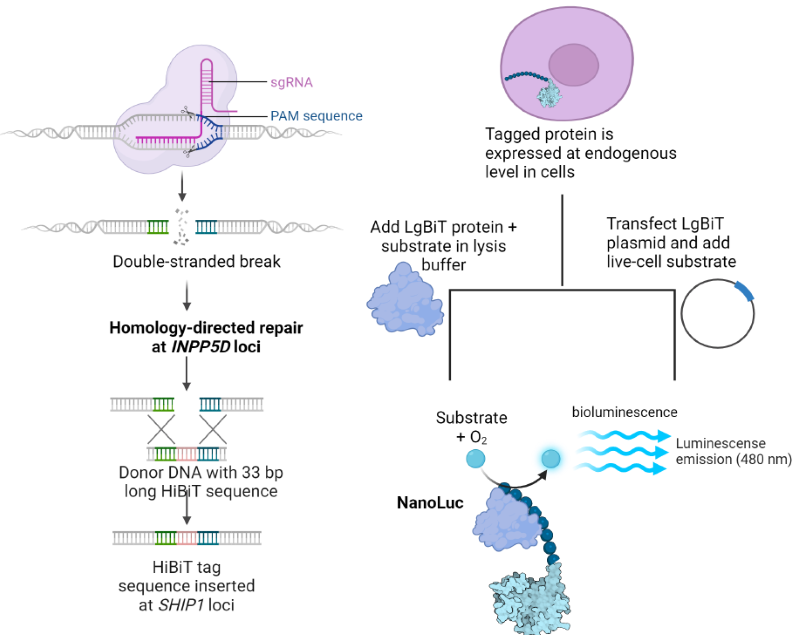
Figure 3. HiBiT LgBiT protein complementary luminescent assay. (A) CRISPR knock-in of HiBiT tag. (B) Live cell or endpoint luminescent assays to monitor SHIP1 protein abundance. Image Credit: Image courtesy of Tika R. Malla et al., in partnership with ELRIG (UK) Ltd.
Validation of HiBiT-SHIP1 expression
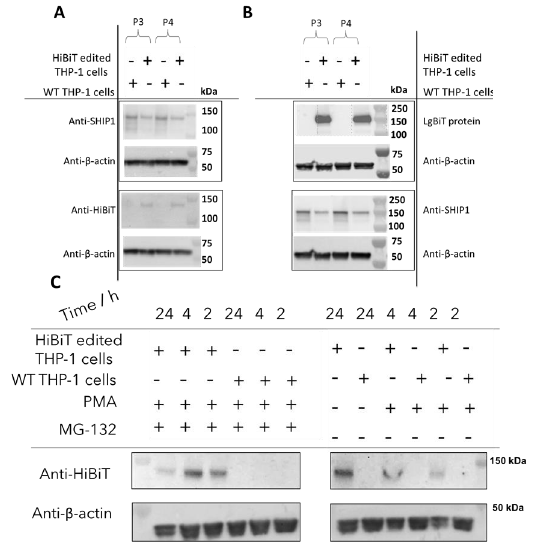
Figure 4. Validation of expression of HiBiT tagged SHIP 1 in CRISPR edited THP 1 cell line using (A) antibody dependent or (B) antibody free HiBiT tag detection (C) HiBiT tagged SHIP 1 is degraded by the proteasomal pathway. Image Credit: Image courtesy of Tika R. Malla et al., in partnership with ELRIG (UK) Ltd.
Optimization of 384 well cell-based assay
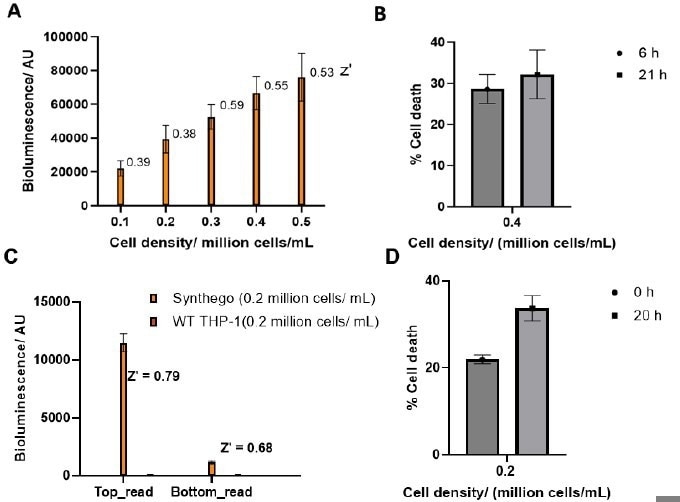
Figure 5. Optimization of multiplexed (A) bioluminescence assay with (B) cell death in a 1536 white plate using an E Clip tip pipette. Optimization of multiplexed (C) bioluminescence assay with (D) cell death in a 384 clear bottom white plate using an E Clip tip pipette. Image Credit: Image courtesy of Tika R. Malla et al., in partnership with ELRIG (UK) Ltd.
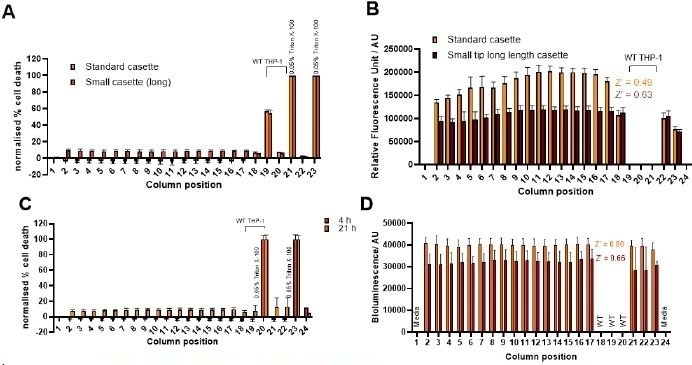
Figure 6. Automation of multiplexed (A, C) cell death and (B,D) bioluminescence assay with different cassettes in multidrop combi (panels A and B) (Thermo Scientific) for high throughput 384 well plate assay. Panels C and D include preincubation of plates for 4 h and 21 h prior to cell toxicity and lytic assay. Image Credit: Image courtesy of Tika R. Malla et al., in partnership with ELRIG (UK) Ltd.
Application of multiplexed complement assay to screen degrader library in 384 well format
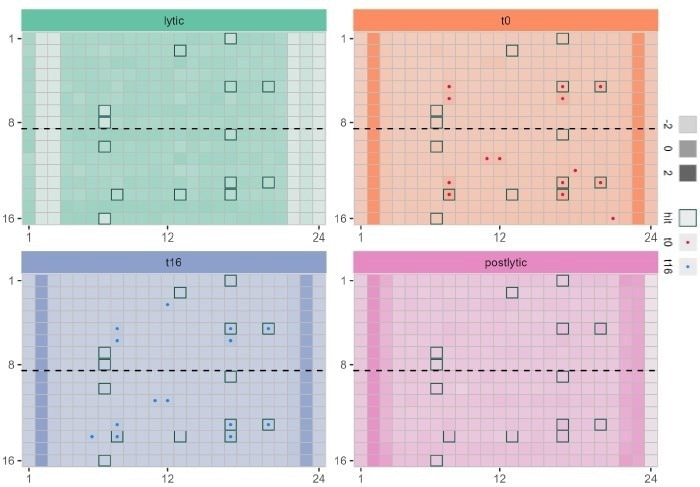
Figure 7. Application of multiplexed bioluminescent assay in 384 well plate to monitor protein degradation (red) compound toxicity immediately after adding to cells (blue) and compound toxicity after incubation with cells for 21 h ..(green) and monitoring the uniformity of cell lysis in all wells (purple) Wells with border indicate top 20 wells with low HiBiT LgBiT bioluminescent signal. Image Credit: Image courtesy of Tika R. Malla et al., in partnership with ELRIG (UK) Ltd.
Application of multiplexed complement assay to screen degrader library in 1536 well format
The assay volume and cell density were optimized to miniaturize the assay in a 1536-well format. Screening with a degrader library in a plate is shown (Figure 8).
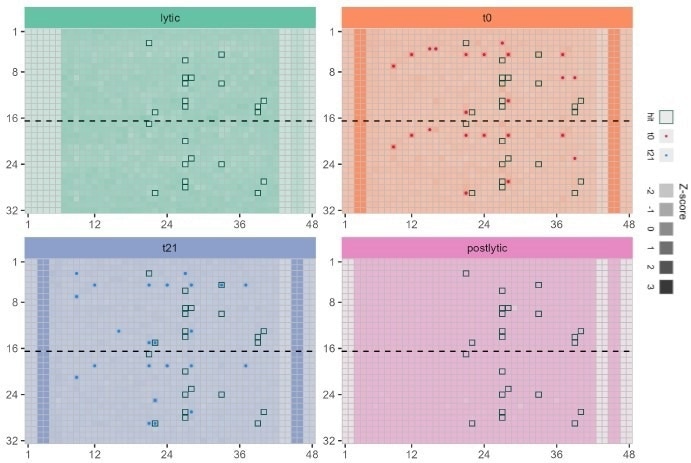
Figure 8. Application of multiplexed bioluminescent assay in 1536 well plate to monitor protein degradation (red) compound toxicity immediately after adding to cells (blue) and compound toxicity after incubation with cells for 21 h ..(green) and monitoring the uniformity of cell lysis in all wells (purple). Image Credit: Image courtesy of Tika R. Malla et al., in partnership with ELRIG (UK) Ltd.
Summary
Validation experiments for HiBiT tagging of edited THP 1 cell lines have successfully been optimized. Immunofluorescence imaging will monitor the localization of WT and HiBiT-tagged SHIP1. A robust high-throughput assay for SHIP1 degradation has been developed in both 384 and 1536 well formats. S screening of a small-molecule compound degrader library has identified preliminary hits, which will be validated by Western Blot and proteomics studies.
References
- Zheng, H., et al. (2018). TREM2 in Alzheimer’s Disease: Microglial Survival and Energy Metabolism. Frontiers in Aging Neuroscience, [online] 10. https://doi.org/10.3389/fnagi.2018.00395.
- Pedicone, C., et al. (2020). Pan-SHIP1/2 inhibitors promote microglia effector functions essential for CNS homeostasis. Journal of cell science, 133(5). https://doi.org/10.1242/jcs.238030.
- Obst, J., et al. (2020). Targeting SHIP1 for therapeutic intervention in Alzheimer’s disease. Alzheimer s & Dementia, 16(S9). https://doi.org/10.1002/alz.045839.
- Bradshaw, W.J., et al. (2024). Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2. Structure. [online] https://doi.org/10.1016/j.str.2024.01.005.
About the University of Oxford
Oxford is an independent and self-governing institution consisting of the University, its divisions, departments and faculties, and the colleges.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devic
Last Updated: Nov 18, 2024