This article is based on a poster originally authored by Daniel A. Barr, Mario Öeren, Peter A. Hunt, Jonathan D. Tyzack, Tomáš Chrien, Tamsin E. Mansley and Matthew D. Segall, which was presented at ELRIG Drug Discovery 2024 in affiliation with Optibrium Limited.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Introduction
Unexpected metabolism can lead to the failure of many late-stage drug candidates or even the withdrawal of approved drugs. Therefore, during early research, it is important to predict the routes, sites, and products of metabolism of potential drug-like molecules. This will help alleviate these risks and aid in identifying metabolites from in vitro/in vivo experiments.
Optibrium’s mechanistic metabolism models cover metabolism by cytochrome P450 (CYP), aldehyde oxidase (AOX), flavin-containing monooxygenase (FMO), UDP-glucuronosyltransferase(UGT), and sulfotransferase (SULT) enzymes.1-5 By combining these models, metabolic pathway analysis proposes the most likely metabolites with greater precision than other methods, assisting in metabolite identification studies and enabling potentially active, reactive, or toxic metabolites to be identified.6
MetabolicInstability
Predicting sites of metabolism can be useful in designing molecules with improved stability, as illustrated in Figure 1. Site lability provides an absolute measure of the susceptibility of each site to metabolism by CYP3A4, represented by the histograms and composite site lability (CSL) illustrated below. Blocking the most labile sites results in significantly improved CYP3A4 half-life.
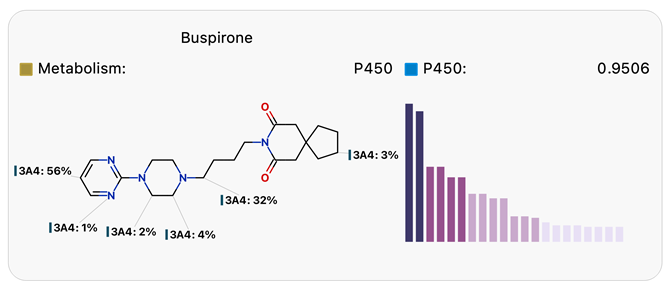
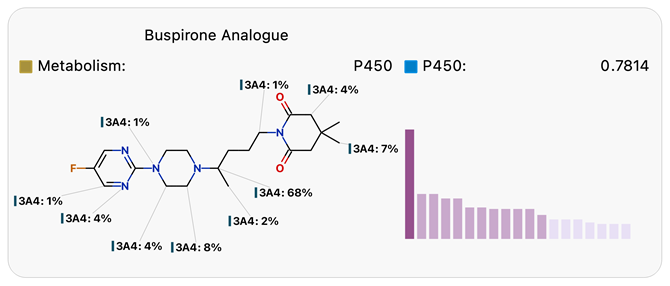
Figure 1. Regioselectivity diagrams and site lability histograms for Buspirone (top) and its more stable analog (bottom) from Tandon.7 Image Credit: Image courtesy of Daniel A. Barr et al., in partnership with ELRIG (UK) Ltd.
Genetic polymorphisms
Genetic polymorphisms in some P450 isoforms, e.g., CYP2D6, CYP2C9, and CYP2C19, significantly increase drug exposure in substantial subpopulations. Metabolism predominantly by a single polymorphic isoform indicates an increased risk, e.g., Metoprolol.8 Predictive modeling using WhichP450™ (Figure 2) identifies the P450 isoforms that are most relevant to consider and enables the prioritization of metabolism phenotyping experiments to confirm the risk identified by the model.
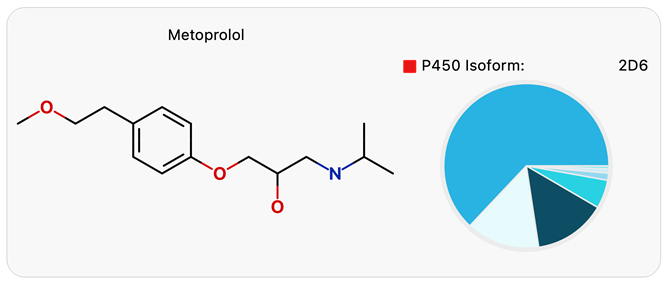
Figure 2. WhichP450 model results for Metoprolol show likely metabolism by CYP2D6. Image Credit: Image courtesy of Daniel A. Barr et al., in partnership with ELRIG (UK) Ltd.
Metabolite prediction
The complex nature of metabolic pathways means there is often a trade-off between precision and sensitivity in metabolite prediction. Predicting every possible metabolite using Optibrium’s Phase I and Phase II models finds all known metabolites (Figure 3, left) but with unacceptably low precision. Combining the metabolism models with trained heuristics to focus on the most likely metabolites increases the precision by an order of magnitude while retaining good sensitivity (Figure 3, right).
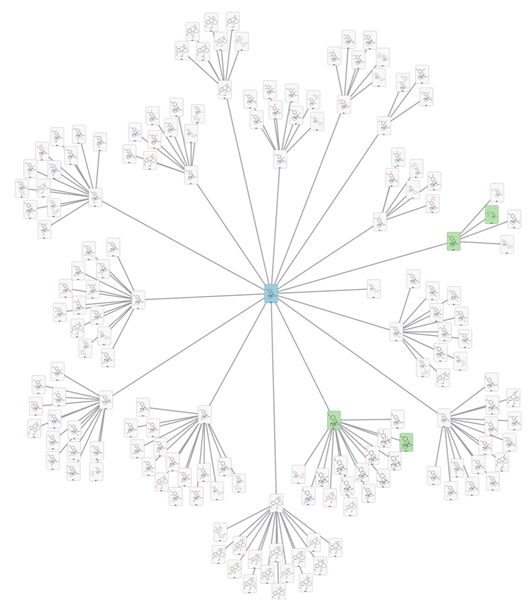
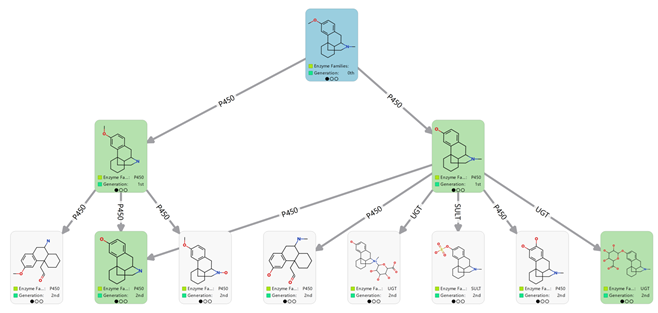
Figure 3. Predicted human metabolism network for dextromethorphan (cyan card), before (top) and after (bottom) applying heuristics to assess the most likely metabolites. Heuristics increased the precision from 4% to 40% without impacting the sensitivity of 100%. Known experimental metabolites are shown in green cards. Image Credit: Image courtesy of Daniel A. Barr et al., in partnership with ELRIG (UK) Ltd.
Preclinical insights
Focusing on microsomal stability can lead to unexpected metabolism by other enzymes in late-stage studies. Falnidamol is an example of a clinical failure due to poor oral bioavailability caused by unexpected metabolism by AOX, which was missed in preclinical studies using rats and dogs.9 The WhichEnzyme™ model6 identifies AOX as most relevant to consider for Falnidamol’s metabolism (Figure 4) and enables prioritization of appropriate in vitro and PK studies; using guinea pigs or rhesus monkeys in preclinical studies would have revealed the risk of metabolism by AOX.
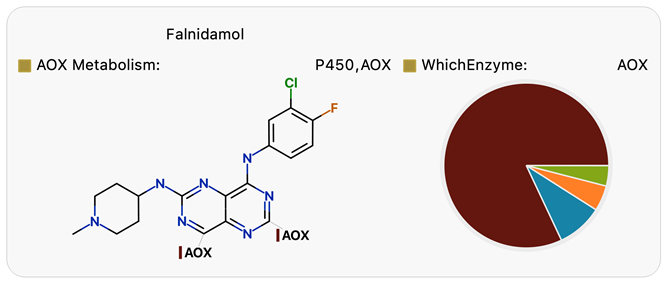
Figure 4. WhichEnzyme model results for Falnidamol indicate that AOX is likely to be an important route of metabolism. Image Credit: Image courtesy of Daniel A. Barr et al., in partnership with ELRIG (UK) Ltd.
Drug-drug interactions
Compounds metabolized by only a single isoform of a single enzyme pose a significant risk for drug-drug interactions (DDIs). Inhibition or induction of that isoform by a co-administered drug can lead to significant changes in exposure. Optibrium’s WhichEnzyme and WhichP450 reveal that Simvastatin is primarily metabolized by CYP3A4 (Figure 5) and that coadministration with potent CYP3A4 inhibitors such as Ketoconazole is contraindicated. These predictions are consistent with experimental results10 and provide insights to guide phenotyping experiments and to evaluate drug combinations.
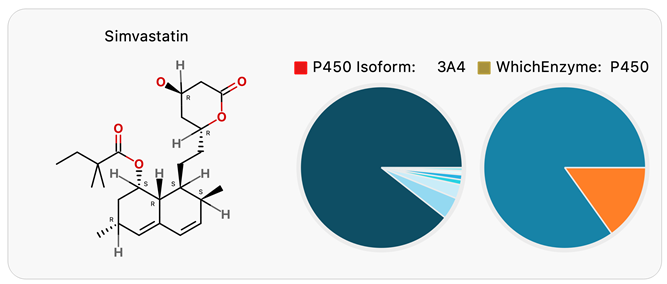
Figure 5. The WhichEnzyme and WhichP450 models for Simvastatin indicate a high risk of drug-drug interactions. Image Credit: Image courtesy of Daniel A. Barr et al., in partnership with ELRIG (UK) Ltd.
References
- Öeren, M., et al. (2022). Predicting Regioselectivity of AO, CYP, FMO, and UGT Metabolism Using Quantum Mechanical Simulations and Machine Learning. 65(20), pp.14066–14081. https://doi.org/10.1021/acs.jmedchem.2c01303.
- Öeren, M., et al. (2023). Predicting Regioselectivity of Cytosolic Sulfotransferase Metabolism for Drugs. 63(11), pp.3340–3349. https://doi.org/10.1021/acs.jcim.3c00275.
- Öeren, M., et al. (2020). Predicting reactivity to drug metabolism: beyond P450s—modelling FMOs and UGTs. Journal of Computer-Aided Molecular Design, 35(4), pp.541–555. https://doi.org/10.1007/s10822-020-00321-1.
- Tyzack, J.D., Hunt, P. and Segall, M.D. (2016). Predicting Regioselectivity and Lability of Cytochrome P450 Metabolism Using Quantum Mechanical Simulations. Journal of Chemical Information and Modeling, 56(11), pp.2180–2193. https://doi.org/10.1021/acs.jcim.6b00233.
- Hunt, P., Segall, M.D. and Tyzack, J.D. (2018). WhichP450: a multi-class categorical model to predict the major metabolising CYP450 isoform for a compound. Journal of Computer-aided Molecular Design, 32(4), pp.537–546. https://doi.org/10.1007/s10822-018-0107-0.
- Öeren, M., et al. (2023). Predicting routes of phase I and II metabolism based on quantum mechanics and machine learning. Xenobiotica, 1–49. https://doi.org/10.1080/00498254.2023.2284251
- Tandon, V.K., Singh, K.A. and Goswamy, G.K. (2004). 1- And 2-substituted naphthalenes: a new class of potential hypotensive agents. Bioorganic & Medicinal Chemistry Letters, 14(11), pp.2797–2800. https://doi.org/10.1016/j.bmcl.2004.03.080.
- Meloche et al. (2020) . CYP2D6 polymorphism and its impact on the clinical response to metoprolol: A systematic review and meta-analysis. BJCP, 86, 1015-1033. https://doi.org/10.1111/bcp.14247
- Hutzler, J.M., Ring, B.J. and Anderson, S.R. (2015). Low-Turnover Drug Molecules: A Current Challenge for Drug Metabolism Scientists. Drug Metabolism and Disposition, 43(12), pp.1917–1928. https://doi.org/10.1124/dmd.115.066431
- Akram, K. and Pearlman, B.L. (2007). Congestive heart failure-related anemia and a role for erythropoietin. International Journal of Cardiology, 117(3), pp.296–305. https://doi.org/10.1016/j.ijcard.2006.05.071.
About Optibrium Ltd. 
Founded in 2009, Optibrium continues to develop new products and research novel technologies to improve the efficiency and productivity of the drug discovery process. Optibrium works closely with its broad range of customers and collaborators that include leading global pharma, agrochemical and flavoring companies, biotech and academic groups.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 18, 2024