This article is based on a poster originally authored by Mat Calder, Ben Durham, Amy Prosser, Miguel Coelho, Aimee Blair, Jack Cobb, Marta Falcicchio, Ruben Alvarez Fernandez, Emma Ford, Alberto Moreno de la Gandara, Ilaria Giovannelli, Penny Hayward, Sajaana Jeyaseelan, Matt Jones, Ásta-Björk Jonsdottir, Navrohit Kandola, Mick Knaggs, Tabitha Morgan, Kathleen Santos, Poonam Shah, Emma Stanway, Aishwarya Sundaresh, Chloe Tarry, Lucy Walker, Robert Yan, Willie Yen, Benedict Cross, Rich Boyce, and Christian Dillon, which was presented at ELRIG 2024 in affiliation with CN-Bio.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.
The rise of targeted protein degradation (TPD) is rapidly changing perceptions about therapeutic target druggability and is rewriting many preconceived notions on drug design.
Most monovalent molecular glue and heterobifunctional degraders in clinical and preclinical development rely predominantly on recruiting a single E3 ligase, Cereblon. Arising resistance, toxicities, and restricted Protein-of-Interest scope may limit clinical potential, highlighting the need to uncover novel TPD mechanisms.
To expand the potential of this modality, PhoreMost has developed SITESEEKER®, a screening technology operating at substantially greater complexity than extant target discovery platforms. SITESEEKER® utilizes computationally derived encoded mini-protein fragments with huge shape diversity to systematically identify novel degrader mechanisms and define functionally active binding sites on targets.
SITSEEKER® allows for the discovery of targets that may be missed through traditional gene editing or knockdown approaches and can also provide valuable mechanistic insights that help inform, unlock, and truncate the path from target ID to drug discovery.
This article identifies a cache of degrader motifs that showcase the breadth of proteome space yet to be explored within TPD. It also demonstrates how peptide motifs can drive degradation selectively with the potential to be translated into tissue- or cancer-selective degraders.
Using combinatorial screening, the functional dependencies of prioritized degraders have been mapped to their cognate E3 ligase, giving rise to several E3 ligases that could be hijacked for TPD.
PhoreMost is progressing a pipeline of monovalent and heterobifunctional oncology degrader programs arising from its platform.
The study demonstrates the effective hijacking of selected E3 ligases, identified by SITESEEKER®, by discovering high-affinity small-molecule binders and subsequent discovery of efficient heterobifunctional degraders against selected Proteins of Interest (POIs).
SITESEEKER® and targeted protein degradation
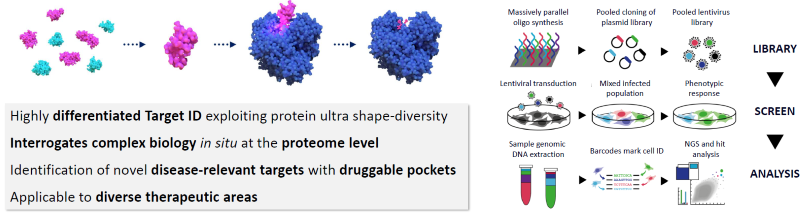
Figure 1. Outline of SITESEEKER® Phenotypic Screening Platform. Image Credit: Zyme Communications Ltd
- PROTACs and Molecular Glues (MGs) are a new class of therapeutic small molecules that hijack the UPS to eliminate disease-causing proteins.
- Most PROTACs/MGs are based on Cereblon-binding IMiD drugs with many known limitations.
- PhoreMost is applying its SITESEEKER® technology to identify and translate novel TPD mechanisms applicable to both heterobifunctional and molecular glue degraders.
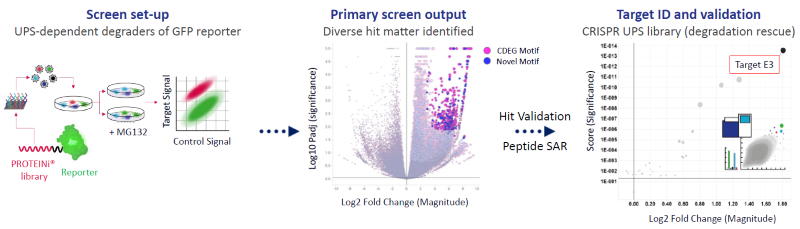
Figure 2. A SITESEEKER® degrader screen identifies 1000s of unique miniprotein sequences driving the degradation of a GFP-reporter. Image Credit: Zyme Communications Ltd
A 300,000 diverse mini-protein (PROTEINi®) library inspired by the human E3 ligase interactome was fused to GFP (left). The screen returned 1000s of diverse hits capable of degrading GFP in a UPS-dependent manner (center). Cognate targets driving GFP degradation of validated PROTEINi® hits were identified by CRISPR (Right) or siRNA perturbation screening (not shown).
Results
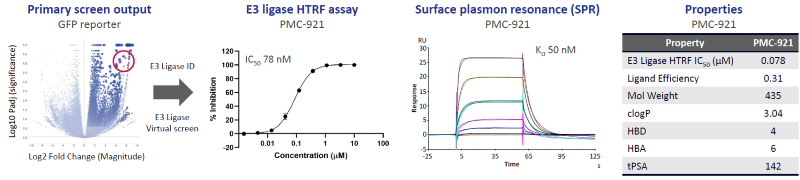
Figure 3. Discovery and optimization of high-affinity small molecule binders to a Cullin-RING E3 Ligase. Image Credit: Zyme Communications Ltd
A Cullin-RING E3 ligase was identified from a cluster of PROTEINi® hits. A virtual screen of 8M compounds and subsequent validation/optimization led to two series of high-affinity small molecule binders (e.g., PMC-921). Ligand binding to the ligase degron pocket and exit vectors for heterobifunctional degrader discovery were validated and confirmed by crystallography (not shown).
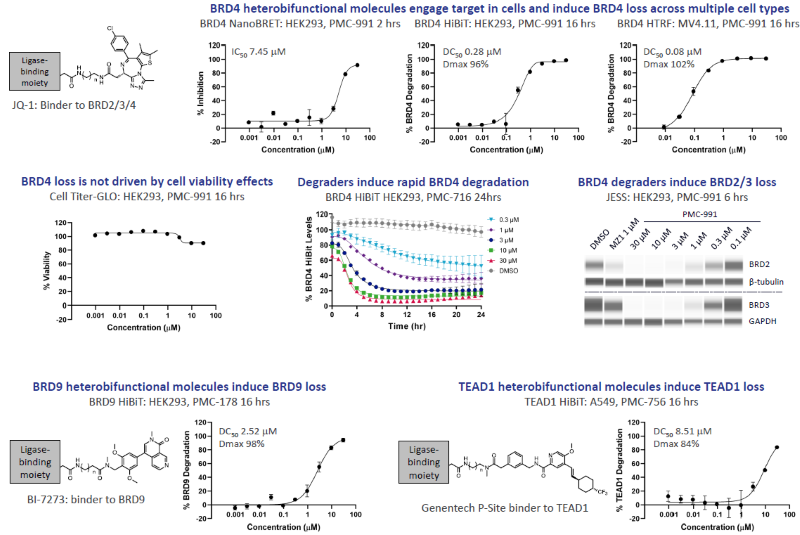
Figure 4. Heterobifunctional small molecules can effectively degrade a diverse range of POIs with rapid degradation onset. Image Credit: Zyme Communications Ltd
BRD4, BRD9, and TEAD1 heterobifunctional molecules were synthesized by exploiting two independent exit vectors and a range of linker lengths/types with the POI binders indicated.
Optimized degraders engaged with the target in cells, leading to ubiquitination (not shown) and rapid degradation in multiple cell models. Effective degradation of a diverse range of POIs was demonstrated.
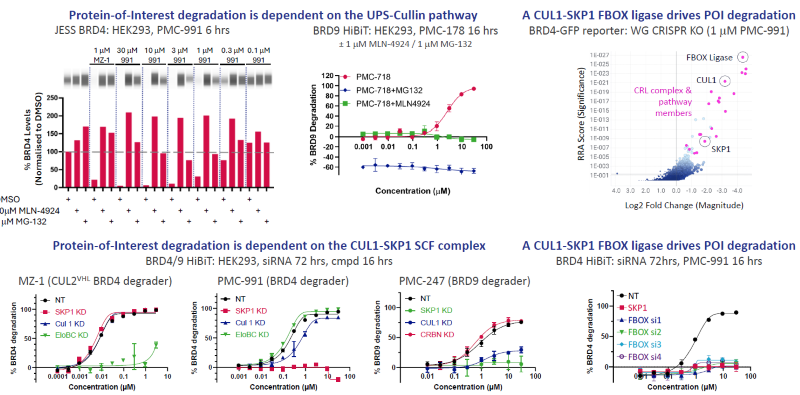
Figure 5. PhoreMost degraders function through a differentiated TPD mechanism of action involving the CUL1-SKP1 SCF complex. Image Credit: Zyme Communications Ltd
POI degradation depends on the Cullin-UPS pathway (MLN-4924/MG-132 rescues degradation). siRNA experiments demonstrated a key functional dependency on the CUL1-SKP1 SCF complex.
Whole-genome CRISPR screen/siRNA studies confirm that a single CUL1-SKP1 substrate receptor, an FBOX E3 ligase, mediates POI degradation.
The FBOX ligase is highly upregulated in many cancers (not shown), raising the possibility of enhanced cancer-selective degradation.
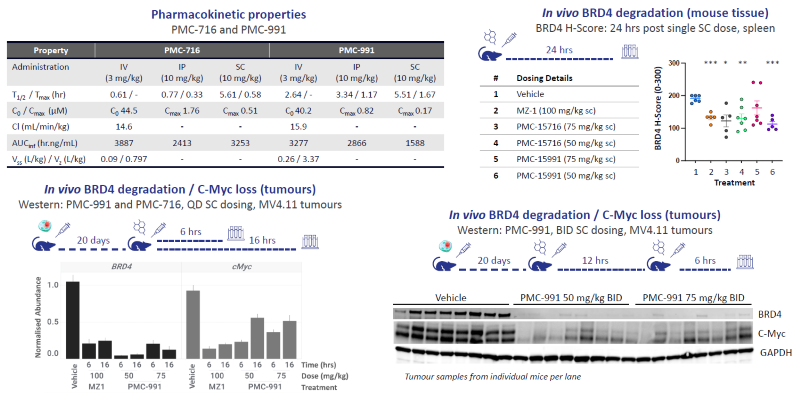
Figure 6. Effective in vivo degradation of BRD4 in mouse tissue and tumors. Image Credit: Zyme Communications Ltd
PK properties of PMC-716 and PMC-991 predicted sufficient target coverage for in vivo BRD4 pharmacodynamic studies. Mice were dosed subcutaneously with tissue or MV4.11 xenografted tumors harvested as indicated.
Significant reductions in BRD4 levels in mouse tissue and BRD4/C-Myc levels in implanted tumors were observed with superior degradation compared to MZ-1. BID dosing leads to complete loss of tumor-associated BRD4 levels.
Conclusions
- SITESEEKER® platform identifies novel mechanisms for TPD and provides enabling tools for target validation and drug discovery.
- High-affinity small molecules were identified, binding an unprecedented Cullin-RING ligase.
- Heterobifunctional molecules can degrade a diverse set of POIs.
- PhoreMost degraders operate through a single FBOX substrate receptor of the CUL1- SKP1 SCF complex, providing a differentiated MOA to VHL/CRBN-based degraders.
- Degraders are highly active in tumor-bearing mouse models.
PhoreMost activities and pipeline
- SITESEEKER® screens focused on new TPD mechanisms and rationalizing MG discovery.
- Pipeline of POI degraders in oncology/inflammation based on its novel TPD platforms.
- PhoreMost has several industry alliances across diverse therapeutic areas.
TPD Platform Discovery, SITESEEKER®. Source: Zyme Communications Ltd
| Target |
Area / Indication |
Stage |
| CUL1-SCF |
Bifunctional degrader platform |
Discovery |
| Undisclosed |
Bifunctional degrader platforms |
Discovery |
| Undisclosed |
Selective TPD ligase ID |
Validation |
| Cereblon |
Rational molecular glue discovery |
Screen |
Degrader pipeline. Source: Zyme Communications Ltd
| Target |
Area / Indication |
Stage |
| Undisclosed |
Key oncogenic pathway |
Discovery |
| Undisclosed |
Inflammatory diseases |
Discovery |
| Undisclosed |
Synthetic lethality, DDR |
Discovery |
| Undisclosed |
Synthetic lethality, epigenetic |
Discovery |
Alliance partnerships

About PhoreMost

PhoreMost is a new-model drug discovery company based in Cambridge, UK: Using its core expertise to open up new ‘druggable’ target space and working with a global network of co-invested academic and industrial collaboration partners, we aim to bring a wide array of novel ‘targeted’ therapies more efficiently to market and pass these cost savings onto patients.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 18, 2024