This article is based on a poster originally authored by Smith Frank, Boute Melodie, Ruez Richard, Amandine Gerstenberg and Ramos Corinne.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Background
Immune checkpoint inhibitors (ICIs) have revolutionized cancer treatment, but many patients with metastatic cancer develop resistance. Understanding the mechanisms behind this resistance is essential for improving treatment outcomes. Traditional techniques struggle to capture the complex spatial interactions within the tumor microenvironment that drive resistance.
Spatial proteomics offers new insights by mapping gene expression and protein activity in the context of tissue architecture. These technologies enable the discovery of region-specific resistance mechanisms and novel therapeutic targets, providing a deeper understanding of how immune cells and tumors interact in resistant metastatic sites.
Introduction
Metastatic castration-resistant prostate cancer (mCRPC) remains a difficult-to-treat condition, often resistant to conventional therapies, including hormone-based treatments. Immunotherapy with checkpoint inhibitors such as CTLA-4 and PD-1 has been explored as a treatment option for these patients. Unfortunately, these therapies have provided little to no clinical benefit. This lack of response is likely due to the complex and unique characteristics of the tumor microenvironment in mCRPC.
In this analysis, we examine the tumor microenvironment mCRPC using spatial analysis to understand the mechanisms behind the resistance to immune checkpoint inhibition. Our goal is to identify new potential therapeutic targets within the tumor microenvironment that could improve the immune system's ability to recognize and destroy cancer cells. Through this exploration, we aim to uncover pathways or cellular interactions that could be disrupted by novel drug therapies, leading to more effective treatment strategies for mCRPC.
This investigation will enhance our understanding of immune evasion in mCRPC and point to new directions for drug development to improve patient outcomes.
Method
Formalin-fixed, paraffin-embedded (FFPE) sections from surgically resected, retrospectively collected metastatic castration-resistant prostate cancer (mCRPC) tissues from 30 patients treated with anti-cancer immunotherapy were analyzed.
Using a high-plex imaging mass cytometry method, the study comprehensively evaluated the tumor and its microenvironment by employing a panel of 30 specific markers (Fig. 1). This dual assessment enabled an in-depth analysis of the immune context within the surrounding microenvironment and cellular changes occurring directly within the tumor tissue itself.
All patients included in the study did not respond to immunotherapies, emphasizing the importance of exploring new therapeutic approaches to target tumor cells and their microenvironmental interactions.
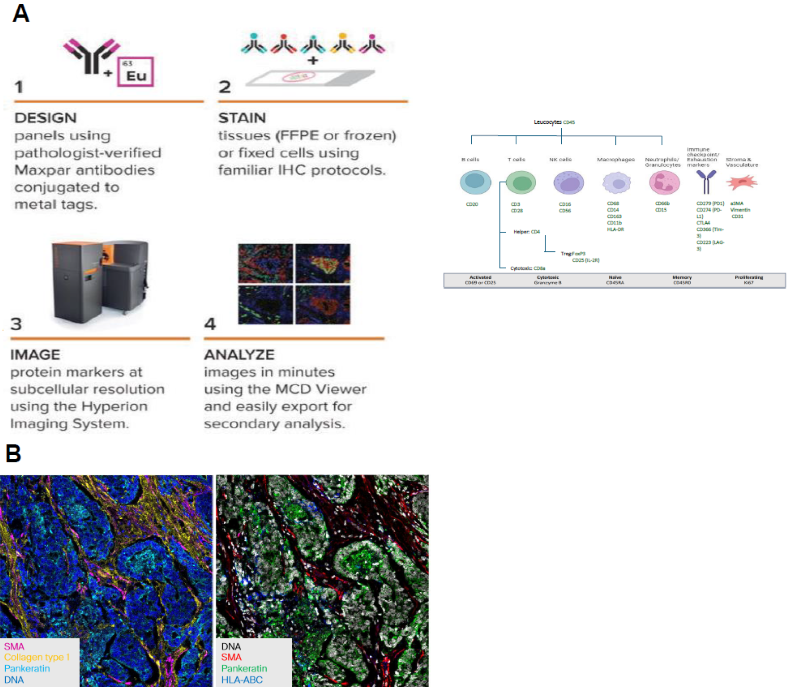
Figure 1. A- Imaging Mass Cytometry high-multiplex workflow. B- High-Plex Immunostaining. Image Credit: Image courtesy of Smith Frank et al., in partnership with ELRIG (UK) Ltd.
Cell analysis workflow
Deep learning was applied for cell segmentation based on the signal from a DNA-binding cell intercalator tagged with natural Iridium isotopes (191 and 193). Subsequently, unsupervised analysis and clustering were performed to group cells according to the markers shown in the table in Figure 1. No predefined categories, such as T cells or B cells, were used.
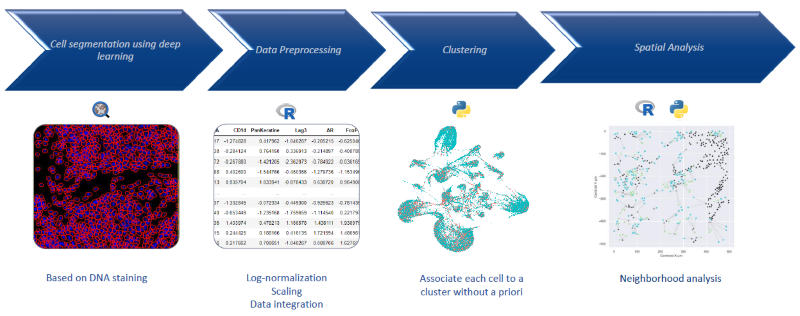
Figure 2. Automatized cell clustering workflow. Image Credit: Image courtesy of Smith Frank et al., in partnership with ELRIG (UK) Ltd.
Results
The analysis of mCRPC tissues revealed key characteristics of the tumor microenvironment. Despite readily detectable CD3+ T cells (Fig. 3), there was a lack of checkpoint protein expression for CTLA-4, PD-1, and Lag3, which are essential for T-cell activation and engagement with tumor antigens. This absence suggests that T cells within these tissues cannot mount an effective anti-tumor response.
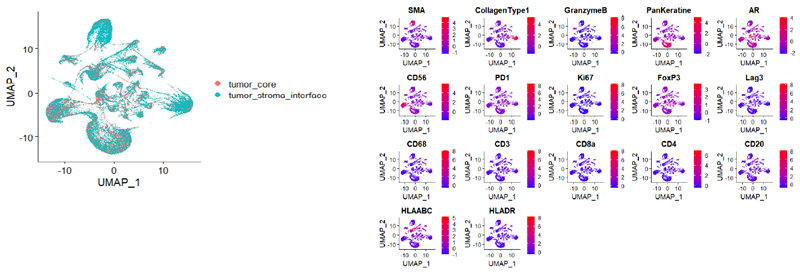
Figure 3. Unsupervised Analysis of mCRPC Tumor Microenvironment Reveals Distinct Immune Subpopulations and Drug Resistance Profiles. Image Credit: Image courtesy of Smith Frank et al., in partnership with ELRIG (UK) Ltd.
The T cells present were found predominantly in a tumor-excluded topology, meaning they were located at the periphery of the tumor rather than infiltrating the core. This spatial arrangement indicates that the T cells are physically separated from tumor cells, reducing their ability to engage with and destroy cancer cells.
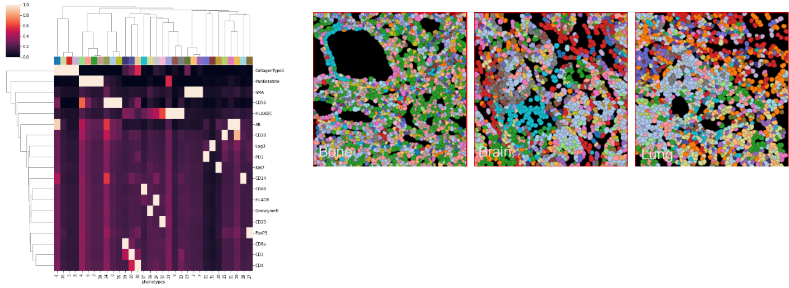
Figure 4. Unsupervised Analysis Shows Checkpoint Loss and T Cell Exclusion in mCRPC. Image Credit: Image courtesy of Smith Frank et al., in partnership with ELRIG (UK) Ltd.
This study suggests an immunosuppressive tumor environment that is not amenable to current checkpoint inhibitor therapies.
Conclusion
Strategies aimed at increasing antigen penetration or modifying the excluded topology to facilitate CD8+ T cell infiltration could potentially overcome cell therapy resistance and enhance the efficacy of immuno-oncology (IO) therapies. By visualizing these tissue clusters at the level of the tumor microenvironment, this unsupervised approach provides valuable insights into the mechanisms of immune exclusion and resistance in mCRPC.
About Aliri Bioanalysis
Aliri is committed to solving industry challenges by bringing innovative bioanalytical and spatial solutions to biotech and pharma, as the complexity of the drug development landscape continues to rise.
Their mission is to support their customers with bringing life-saving therapies to market with speed and agility.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 18, 2024