This article is based on a poster originally authored by Dillon Rinauro, Thomas Peplow, Phil Rawlins, Alec O'Keeffe, Corinne Tovey, Tayo Alleyne-Weir, James Craswell, Kartika Shetty, Michaela Buerdsell, Trevor Askwith and David Cronk, which was presented at ELRIG Drug Discovery 2024 in affiliation with Domainex Ltd.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Introduction
- Accurate measurement of interactions between small molecules/biologics and targets of interest (TOI) is crucial for developing effective therapeutic treatments.
- Surface-based biophysical methods, such as surface plasmon resonance (SPR) and grating-coupled interferometry (GCI), are routinely used to determine the binding kinetics (Fig. 1).
- Though powerful techniques, SPR & GCI can face challenges when quantifying binding interactions, such as when analytes exhibit very slow dissociation rates (long residency times) or when assessing the kinetics of larger, multivalent, complex biologics.
- Methods have been developed to investigate challenging systems by SPR, but it remains to be seen if these can extend to GCI, a more recently developed surface-based technique. We show the development of a fully regenerable GCI surface and streamlined workflows for antibodies/biologics and analytes with long residence times.
- Controlled release of biotinylated substrate from a regenerable GCI surface.

Image Credit: Malvern Panalytical
Controlled release of biotinylated substrate from a regenerable GCI surface
Challenges of current immobilization strategies
- Amine coupling is readily available but heterogeneous and can reduce protein activity.
- NTA affinity capture is possible for his-tagged proteins and regenerable surface, but baseline drift can complicate kinetic analysis.
- Streptavidin capture of biotinylated substrate reduces baseline drift but is not regenerable due to high biotin-streptavidin affinity.
- Non-specific binding of analytes to streptavidin
- Enzymatic site-specific biotinylation of an Avi-tagged protein homogenizes capture and maintains activity.
- Regenerable surfaces can increase screening throughput and reduce consumables costs.
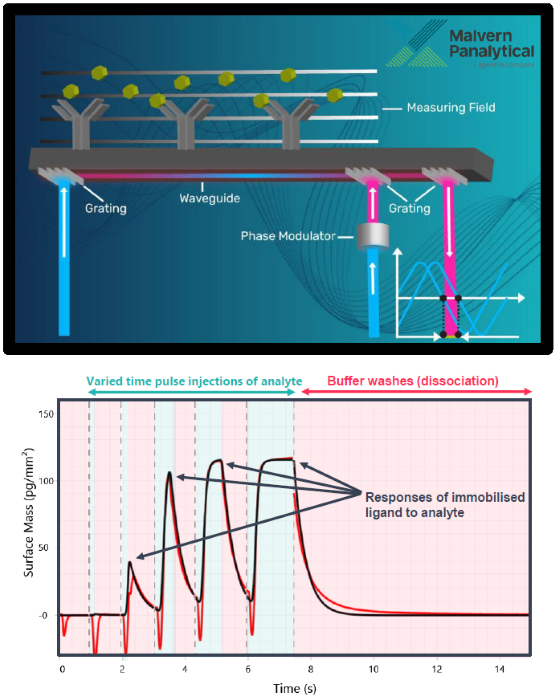
Fig 1. Principles of GCI. Image Credit: Image courtesy of Dillon Rinauro et al., in partnership with ELRIG (UK) Ltd.
Switchavidin
- Engineered for pH-dependent affinity of avidin, with much faster dissociation under slightly acidic conditions
- Further mutated to reduce non-specific binding of analyte
- More flexibility by allowing multiple rounds of immobilization on the same chip
- Improved efficiency of surface-based kinetic studies (Fig. 2)
- At the time of experiments, no established method existed for GCI

Fig 2. General workflow of switchavidin (SwA) capture. Image Credit: Image courtesy of Dillon Rinauro et al., in partnership with ELRIG (UK) Ltd.
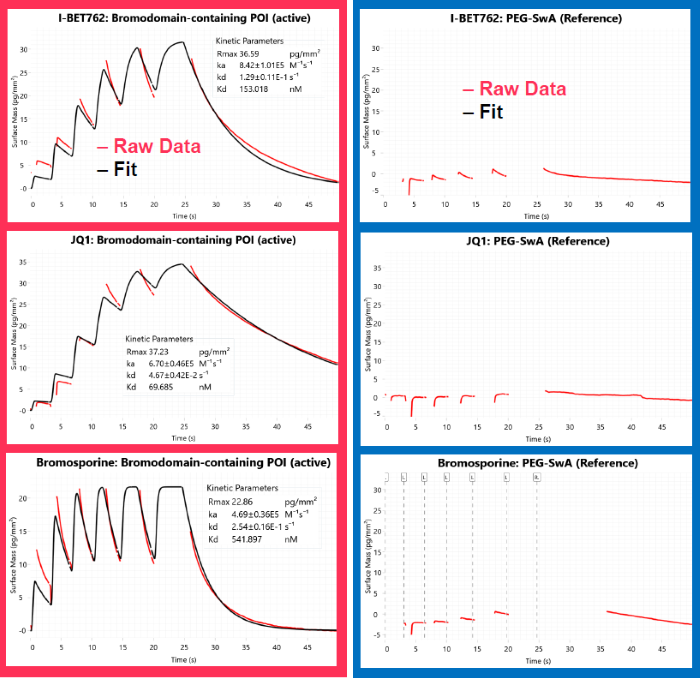
Fig 3a. Method validation of SwA capture using tool compounds. Image Credit: Image courtesy of Dillon Rinauro et al., in partnership with ELRIG (UK) Ltd.
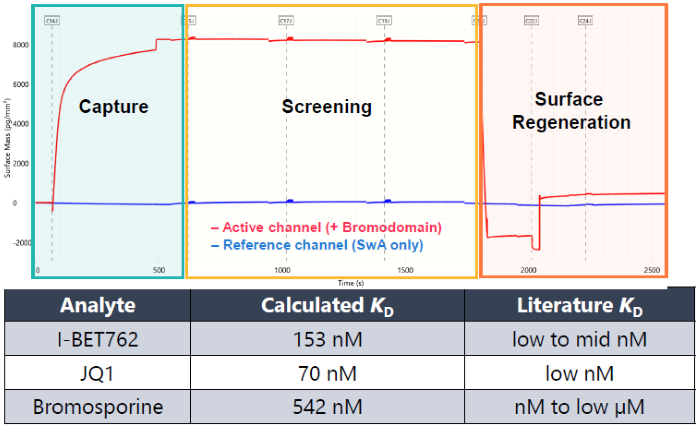
Fig 3b. Measurement of surface density in each stage of workflow. Image Credit: Image courtesy of Dillon Rinauro et al., in partnership with ELRIG (UK) Ltd.
Method validation
- Biotinylated PEG2-amine was covalently attached to a polycarboxylate surface using NHS succinimide cross-coupling
- Biotinylated bromodomains were incubated with Switchavidin
- Three test compounds were screened against bromodomain proteins (Fig. 3a)
- Affinities closely match reported values in literature (Fig. 3b)
- Near complete removal of SwA-bromodomain protein complex using dilute citric acid + SDS
- No surface activity of compounds following chip stripping
Kinetic profiling of antibodies
Challenges
- Multivalent species can bind multiple binding partners, particularly in high-density environments.
- Engaging multiple epitopes simultaneously will enhance the stability of antibody-antigen complexes and increase residence time. This phenomenon is known as avidity (Fig. 4a)
- Avidity can be avoided by capturing the antibody instead of the antigen
- Poor covalent immobilization of antibodies can occlude the Complementarity-determining regions (CDRs) from binding partners (Fig. 4b)
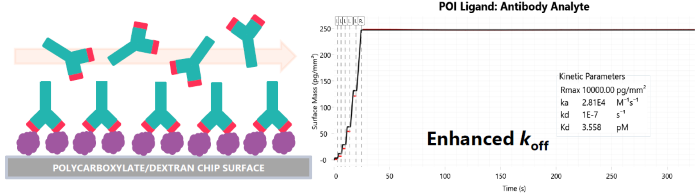
Fig 4a. Avidity effects with antigen immobilization. Image Credit: Image courtesy of Dillon Rinauro et al., in partnership with ELRIG (UK) Ltd.
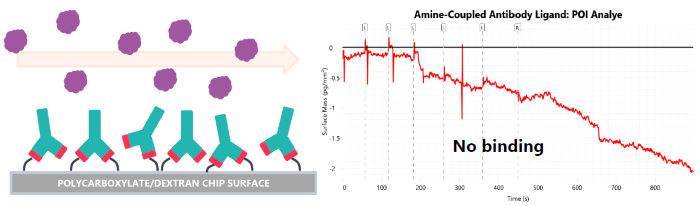
Fig 4b. Poor antibody immobilization strategy. Image Credit: Image courtesy of Dillon Rinauro et al., in partnership with ELRIG (UK) Ltd.
Proteins A & G (PAG)
- Strong affinity for non-binding (Fc region) of antibodies – typically nM
- Covalent immobilization of PAG can properly orient antibodies to measure 1:1 binding events between paratope & antigen
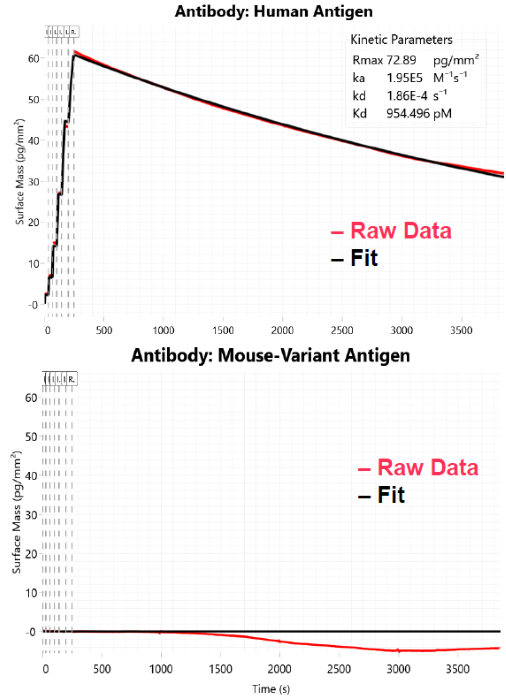
Fig 5. Antibody selectivity after PAG capture. Image Credit: Image courtesy of Dillon Rinauro et al., in partnership with ELRIG (UK) Ltd.
Key findings
- PAG capture of antibody minimizes avidity & rebinding effects; correct orientation of paratope
- pM to nM affinity of antibodies for human variant antigen (Fig. 5)
- Antibody selective for human variant antigen; no binding observed for mouse variant antigen
- Low immunoresponse to alt. mammalian variant antigens may predict performance in pre-clinical studies
Chaser assays for a slow koff
Challenges
- Rebinding of a compound can artificially enhance the koff
- Extended dissociation times require syringe re-filling & re-injection
- Re-injections of buffer introduce noise (Fig. 6) during the wash (dissociation) phase, further decreasing confidence in koff calculations
- koff may be more accurately determined by measuring the return of surface activity using a control/tool compound
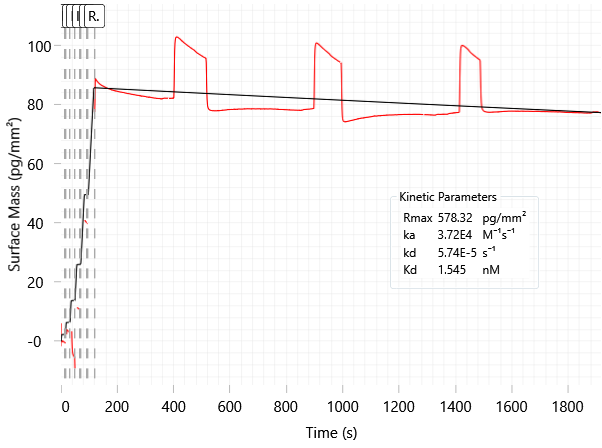
Fig 6. Re-injection spikes observed during long wash phases. Image Credit: Image courtesy of Dillon Rinauro et al., in partnership with ELRIG (UK) Ltd.
Principle workflow
- Protein captured to the chip surface
- The Rmax of a tool compound with a faster koff is recorded at t0
- Protein saturated with slow dissociation (koff) analyte (compound A)
- The tool compound is injected at regular intervals (tn) over a precalculated time window
- As compound A dissociates, more protein is available for the tool compound to bind to
- Rmax will increase over time window
- An Rmax correction is applied: Rmax (t0) – Rmax (tn)
- Half-life calculated using an exponential decay model (Fig. 7)
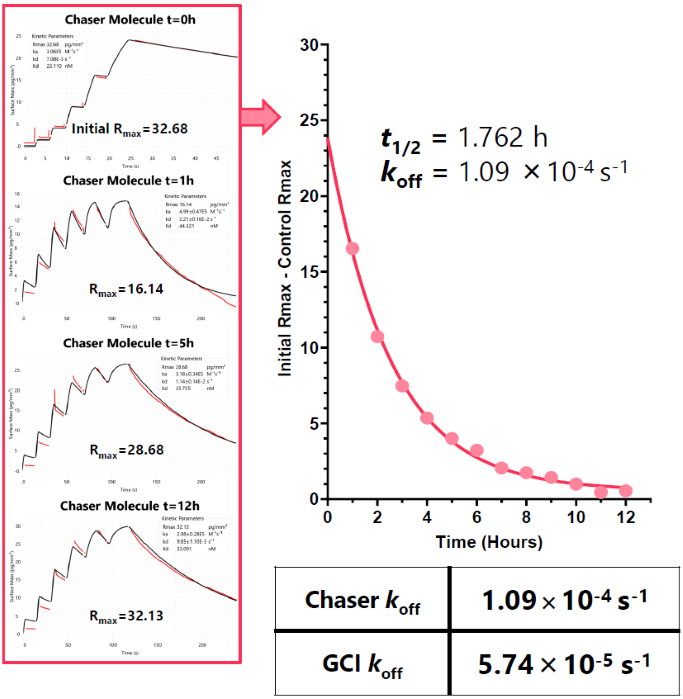
Fig 7. Comparison of chaser assay to a traditional injection mode. Image Credit: Image courtesy of Dillon Rinauro et al., in partnership with ELRIG (UK) Ltd.
Key findings
- Careful selection of tool (chaser) compound required for successful execution of assay
- Chaser assay more accurately calculates dissociation rate constants by reducing re-binding effects for compounds with long half-lives.
- Injection spike interference due to syringe refills eliminated
Summary
- We have developed a fully regenerable GCI chip for biotinylated molecules, whereby their controlled release is regulated via the pH-dependent affinity of an engineered avidin, switchavidin. This increases screening throughput and cost-effectiveness for our clients.
- Proteins A & G (PAG) capture of the Fc region correctly orients the antibody and avoids acidic conditions required for efficient amine cross-coupling. This maintains antibody activity and reduces avidity & rebinding effects to more accurately binding kinetics via a 1:1 model.
- A chaser can be used to more accurately determine the dissociation rate constant of a lead candidate small molecule by reducing re-binding and eliminating re-injection spikes from syringe refills.
About Domainex
Domainex is a fully integrated drug discovery CRO based near Cambridge, UK. It serves pharmaceutical, biotechnology, academic and patient foundations globally. Domainex’s drug discovery service business was established in 2001 and since that time has continued to expand to serve a wider range of international clients including UCB, FORMA Therapeutics, St George’s University, and The Institute of Cancer Research.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 18, 2024