Histones are proteins that condense and package DNA neatly into chromosomes. Modifications to these proteins affect different processes in the cell such as the activation/inactivation of transcription, chromosome packaging, DNA damage and DNA repair. The modification of histones is an important post-translational process that plays a key role in gene expression. The modifications impact this gene expression by changing the structure of chromatin or through recruitment of histone modifiers.
Histones pack DNA into structures called nucleosomes, to fit the DNA molecule into the nucleus. Each of these nucleosomes has two subunits, each comprising the core histones H2A, H2B, H3 and H4, and a linker histone called H1 that acts as stabilizer. The following sections briefly discuss three important types of histone modification.
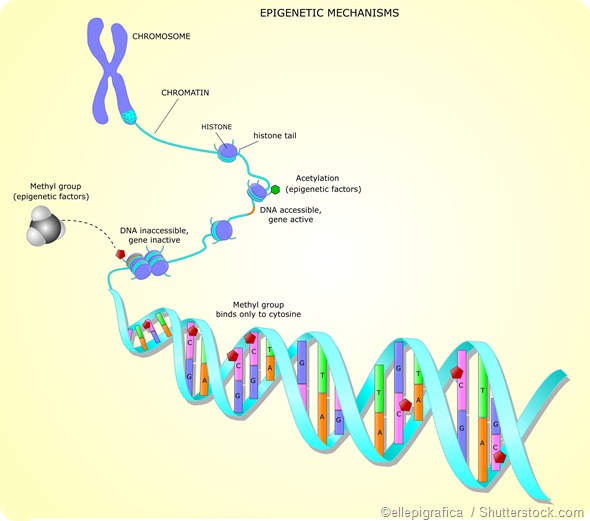
Histone acetylation
Histone acetylation is widely associated with a chromatin structure that is open and therefore accessible to transcription factors, which increases gene expression. It is largely targeted to gene promoter regions. For example, usually, acetylation on H3 is associated with active gene enhancers and promoters.
The enzymes responsible for regulating the acetylation of histone tails are histone acetyltransferases (HATs) and histone deacetylases (HDACs). The preferred targets for HATs are the lysine residues in the tails of histone H3 and H4. Histone acetylation is a reversible reaction. HDACs enable acetyl groups to be removed from histone lysine residues.
An imbalance in histone acetylation has been linked to tumor formation and cancer progression. Being able to determine whether H3 is acetylated at the lysine residue would be useful for characterizing patterns of acetylation and help researchers understand how gene expression is regulated by epigenetics, as well as aiding the development of HAT-targeted drugs.
Histone methylation
Histone methylation is the transfer from S-adenosyl-L-methionine of one to three methyl groups, to lysine or arginine residues of histone proteins. The transfer is catalyzed by histone methyltransferases (HMTs). Depending on the methylation site, the methylation of lysine in H3 and H4 is implicated in both the activation and repression of transcription, while arginine methylation is implicated only in activation of the transcription. The residues are methylated with the help of enzymes called lysine methyltransferases. Functional diversity to the individual methylation sites is provided by the fact that lysine can be mono-, di- or trimethylated.
For instance, both mono- and tri-methylation of K4 on histone H3 cause transcriptional activation, but the former occurs at transcriptional enhancers and the latter occurs at gene promoters. The tri-methylation of K9 and K27 on histone H3 are both a signal for the repression of transcription. Trimethylation of K27 mainly occurs at promoters in gene-rich chromosomal regions and controls developmental regulators in the stem cells of embryos, while trimethylation of K9 usually occurs in gene poor regions such as telomeres or satellite repeats.
Histone phosphorylation
Phosphorylation of core histones is crucial for chromosome condensation while cell division, DNA repair and transcriptional regulation are occurring. Histone kinases add a phosphate group taken from ATP to the hydroxyl group of a target amino-acid side chain. Histone phosphorylation promotes interactions between other histone modifications and provides a platform for effector proteins.
Histone phosphorylation at H2A on T120 and H3 at S10 are required for the compaction of chromatin and regulation of its structure and function during mitosis.
Histone Modification
References
- http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3193420/
- http://www.whatisepigenetics.com/histone-modifications/
- http://www.abcam.com/epigenetics/histone-modifications-a-guide
Further Reading
Last Updated: Feb 26, 2019