Influenza virus is a common cause of human respiratory infection with a high rate of morbidity and mortality, particularly in the elderly and in infants.
 Credit: Katryna Kon/Shutterstock.com
Credit: Katryna Kon/Shutterstock.com
Influenza A belongs to the Orthomyxoviridae family. It has a negative sense RNA genome encoding 11 viral genes, contained within a viral envelope. The viral genes are as follows:
- hemagglutinin (HA)
- neuraminidase (NP)
- matrix 1 (M1)
- matrix 2 (M2)
- nucleoprotein (NP)
- non-structural protein (NSP1)
- non-structural protein 2/nuclear export protein (NS2/NEP)
- polymerase acidic protein (PA)
- polymerase basic protein 1 (PB1)
- polymerase basic protein 2 (PB2)
- polymerase basic protein 1-F2 (PB1-F2)
Outer virus particle
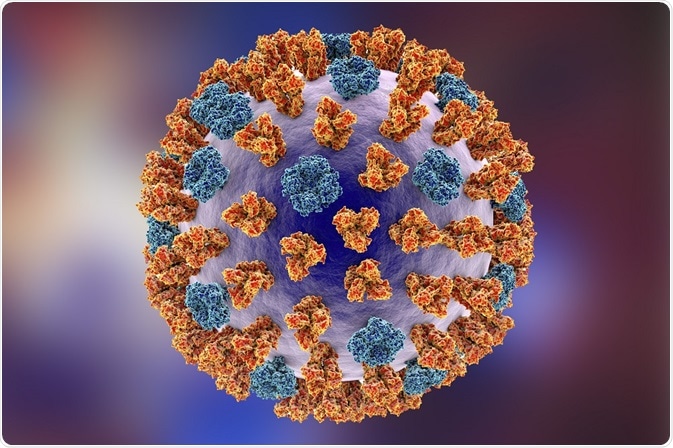
The influenza particle, or virion, is typically spherical, but sometimes filamentous. It has an outer lipid membrane layer called an envelope derived from the host cell that it replicated in. The envelope is covered with glycoproteins HA and NA which form structures like spikes. The ratio of HA to NA molecules is about four to one.
The matrix ion channel M2 penetrates the envelope. The matrix protein M1 is found beneath the lipid membrane. It forms a shell, providing strength and rigidity to the virion. The spherical form influenza A viruses are typically about 100 nm in diameter and filamentous forms can be longer than 300 nm.
Entry into the host cell begins when spikes formed by HA on the surface of the virion bind to sialic acid on the surface of the host cell. The binding of HA to sialic acid is highly species specific. Humans have a different specificity from horses and birds. However, swine viruses recognize both, which makes swine an effective mixing vessel for avian, equine, and human influenza A virus.
Interior of the virus
Inside the viral particle are the 8 viral RNAs that comprise the genome. Each RNA segment is is joined with proteins B1, PB2, PA, and NP. The protein NEP is also found in the interior of the virion. The ribonucleoprotein (RNP) complex within the virion consists of viral RNA segments coated with NP and heterotrimeric RNA-dependent RNA polymerase, which has subunits PB1, PB2 and PA.
Genome structure
The influenza A genome is comprised of eight negative sense, single-stranded viral RNA segments. They are numbered in order of decreasing length. Segments 1, 3, 4, and 5 encode a single protein per segment. Those are PB2, PA, HA, and NP, respectively. Segment 2 contains PB1.
Some strains of influenza A may also code for PB1-F2, a small protein with pro-apoptotic activity, in an alternate reading frame. Segment 6 of influenza A encodes NA protein and in an alternate reading frame, NB matrix protein. Segment 7 codes for M1 matrix protein and through RNA splicing, M2 ion channel. Segment 8 encodes the NS1 protein, and, through mRNA splicing, NS2/NEP.
Each vRNA segment forms a helical hairpin in shape, and is bound by the heterotrimeric RNA polymerase complex. The rest of it is coated with NP. Each segment has non-coding regions at each end of varying lengths. However, the very end of all segments is a highly conserved sequence which serves as a promoter for vRNA replication and transcription. The non-coding regions also contain the mRNA polyadenylation signal and some signals for virus assembly.
The segmentation of the influenza A genome allows the phenomenon of antigenic shift, wherein the virus acquires an HA segment from another influenza subtype. This occurs in cells infected with different viruses, and the resulting virus may encode a wholly novel protein to which humans have no pre-existing immunity.
Influenza animation - flu virus mechanism
Further Reading
Last Updated: Aug 23, 2018