Modifications of the chromatin are crucial for the regulation of nearly all DNA-templated biological processes. Single-cell chromatin modification profiling can be used to assess chromatin modifications, which can provide relationships between epigenetic changes and physiological changes.
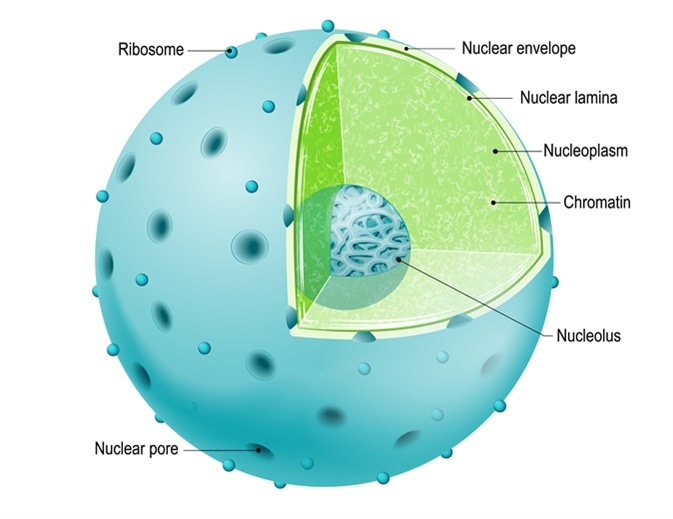
Structure of Nucleus. parts of the cell: nuclear envelope, nucleoplasm, nuclear matrix, chromatin and nucleolus. Image Credit: Designua / Shutterstock
Chromatin Modifications
Chromatin modification is achieved by covalent histone modifications and nucleosome modifications. Histone acetylation, deacetylation, methylation, and phosphorylation are forms of covalent histone modifications, performed by enzymes such as histone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyltransferases (HNMT), and kinases. DNA methylation performed by DNA methyltransferases (DNMT) and DNA phosphorylation performed by kinases are forms of nucleosome modifications. Chromatin remodeling is important to regulate gene expression and other regulatory processes, such as egg cell DNA replication and repair, apoptosis, and chromosome segregation.
Types of Chromatin Modification Profiling
Chromosome immunoprecipitation sequencing
Chromosome immunoprecipitation sequencing (ChIP-seq) can analyze protein interactions with DNA and provide information regarding epigenetic changes. ChIP-seq involves cross-linking protein to DNA and then DNA fragmentation by sonicating. Bead-attached antibodies are then added to immunoprecipitate the target protein.
Chromatin immunoprecipitation quantitative PCR analysis
Chromatin immunoprecipitation quantitative PCR analysis (ChIP-qPCR) can be used to assess the protein-DNA interaction at known genomic binding sites. qPCR primers can be designed against potential regulatory regions (e.g. promoters) when the sites are not known. ChIP-qPCR has a very similar procedure to standard ChIP-qPCR, but at the end of a ChIP-qPCR experiment the resulting DNA undergoes a qPCR and is then analyzed.
Mass cytometry
Mass cytometry combines flow cytometry and mass spectrometry, which is used to discern the properties of cells. Antibodies are conjugated with pure elements and used to label cellular proteins. Cells are then nebulized and sent through an argon plasma which ionizes the antibodies. The metal signals produced are analyzed by a mass spectrometer. This method is used to analyze chromatin modifications.
Discoveries Made Using Chromatin Modification Pprofiling
Aging
Recently, increased amount of epigenetic variations have been observed with aging. Multiplexed mass cytometry analysis is used to profile the levels of chromatin modifications in primary human immune cells at the single-cell level. Differential analysis between younger and older adults was performed, which showed that aging shows an increased heterogeneity between individuals and increased cell-to-cell variability in chromatin modifications. These findings show the impact of age on chromatin modifications.
Breast cancer
Epigenetic changes have been found during malignant transformation in a breast cancer model. The breast cancer model was made by expressing three oncogenes in breast epithelial cells. ChIP analysis was performed, followed by next-generation sequencing of histone methylation. Western blot, quantitative reverse-transcriptase PCR (qRT-PCR), and immunostaining were used to determine the gene expression. The study discovered that two histone marks, involved in the repression of certain oncogenes, were decreased in the breast cancer model.
Immunosuppression
SUMOylation is a post-transcriptional modification involved in various cellular processes such as transcriptional regulation, apoptosis, protein stability, and cell cycle progression. Humans have 3 SUMO proteins (SUMO-1 and SUMO-2/3). A study discovered that herpes viruses (Kaposi’s sarcoma-associated herpesvirus (KSHV)) have evolved mechanisms that directly and indirectly modulate SUMO machinery to avoid the host immune system. The ChIP-seq experiment performed discovered a significant increase in SUMO-2/3 with KSHV reactivation. Gene ontology analysis discovered genes involved in cellular immune responses and cytokine signaling. The KSHV avoids the immune system by increasing the transcription of SUMO-2/3 to hinder an immune response.
Conclusion
Profiling of chromatin modifications modifications allows us to better understand certain biological process and diseases. Further research will provide more knowledge regarding beneficial chromatin changes.
Chromatin Dynamics and Single cell Analysis - Jane Mellor
Further Reading
Last Updated: Aug 23, 2018