RNA (ribonucleic acid) editing is a post-transcriptional alteration of RNA sequences and structures via modification, deletion or insertion of nucleotides. This process has been detected in eukaryotes ranging from single-celled protozoa to plants and mammals; as a result, functionally distinctive proteins can be processed from a single gene.
RNA editing occurs concurrently with transcription and splicing processes in the nucleus or mitochondria. It comes in a myriad of different natures, with all of the diverse editing types distributed intermittently across the phylogenetic spectrum. Two separate types of RNA editing in mammals will be further discussed.
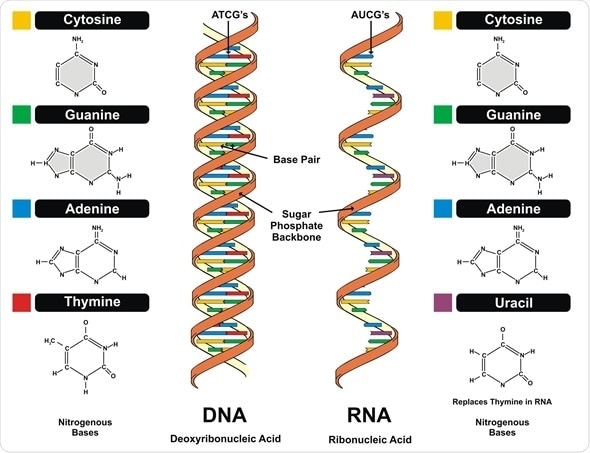
Image Copyright: udaix, Image ID: 108466967 via Shutterstock.com
A-to-I RNA editing
RNA editing by adenosine-to-inosine (A-to-I) base modification successfully generates RNA and protein diversity in higher eukaryotes, selectively reshaping coding and noncoding sequences in nuclear transcripts. The enzymes responsible for this type of editing are known as ADARs (adenosine deaminases acting on RNA), which are pervasive in mammals.
Chemical conversion of adenosine to inosine occurs through removal of an amino group. Prior to editing adenosine hydrogen bonds with thymine, whereas after editing inosine hydrogen bonds with cytosine. A-to-I RNA editing often results in codon changes that alters the function of a chosen protein.
Most of ADAR editing events take place in non-coding regions of the human genome, and current estimates predict that there are more than 40 thousand editing sites in the human transcriptome. Messenger RNAs edited within coding regions at specific adenosines are only known to occur for approximately 30 human genes. Therefore the transcript diversity generated by RNA editing suitably affects regulatory elements in the messenger RNA.
C-to-U RNA editing
A second important type of RNA editing in mammals also relies on base modification and it takes place in apoB (apoprotein B). This process has significant effects on the metabolism of lipoproteins, and it necessitates a single-strand template with clear-cut characteristics in the vicinity of the edited base.
Recently, APOBEC family of enzymes has been discovered that has the ability to deaminate cytidines to uridines on both RNA and DNA in order to mediate this type of RNA editing. This type of RNA editing results in the modification of a glutamine codon to a stop codon and the subsequent synthesis of APOB48 (which is a shorter isoform of APOB100).
APOBEC and C-to-U RNA editing also has an important role in the adaptive immune response, which utilizes antibodies in order to neutralize invading pathogens. Editing of antibody-encoding genes is carried out by an enzyme known as AID (activation-induced cytidine deaminase), also from APOBEC family.
RNA-editing databases
RNA editing sites are already mapped on the reference human genome, and databases that supply all the available and published data linked to RNA editing can be found online. It is even possible to acquire RNA editing information for a specific RNA molecule or gene using matched accession numbers in RNA editing libraries.
Such endeavors already resulted in successful collaborations. For example, Thermo Fisher Scientific, which is one of the leading instrument and reagent providers, partnered with AstraZeneca and provided them with RNA-guide libraries that target individual gene families and human genes in order identify new biological targets.
Thermo Fisher Scientific is also trying to hasten the access to forefront genome-editing applications by employing RNA editing and other genome-modifying techniques. Enabling more pertinent and applicable disease models will improve identification and editing of the target, which can in turn be translated to therapeutics.
AMS Biotechnology or AMSBIO, a company founded in 1987, is now one of the leaders in the field of genome editing, and also provides tissue nucleic acid and protein products for that purpose. Other commercial companies are increasingly joining the quest for RNA-based therapeutics and applications.
Sources
- http://www.jbc.org/content/278/3/1395.full
- http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3086428/
- http://bioinformatics.oxfordjournals.org/content/26/14/1772.long
- www.columbia.edu/.../RNAediting-maas.pdf
- Elliott D, Ladomery M. Molecular Biology of RNA, Second Edition. Oxford University Press, 2016; pp. 268-294.
- Hundley HA. Regulation of Gene Expression Through Inosine-containing Untranslated Regions. In: Mass S, editor. RNA Editing: Current Research and Future Trends. Caister Academic Press, Norfolk, UK, 2013; pp. 193-206.
Further Reading
Last Updated: Aug 23, 2018