Phylogeny is the representation of the evolutionary history and relationships between groups of organisms.
The results are represented in a phylogenetic tree that provides a visual output of relationships based on shared or divergent physical and genetic characteristics.
Phylogenetic analysis is dependent on the type of data inputted, the number of species and the range of evolutionary relationships interpreted.
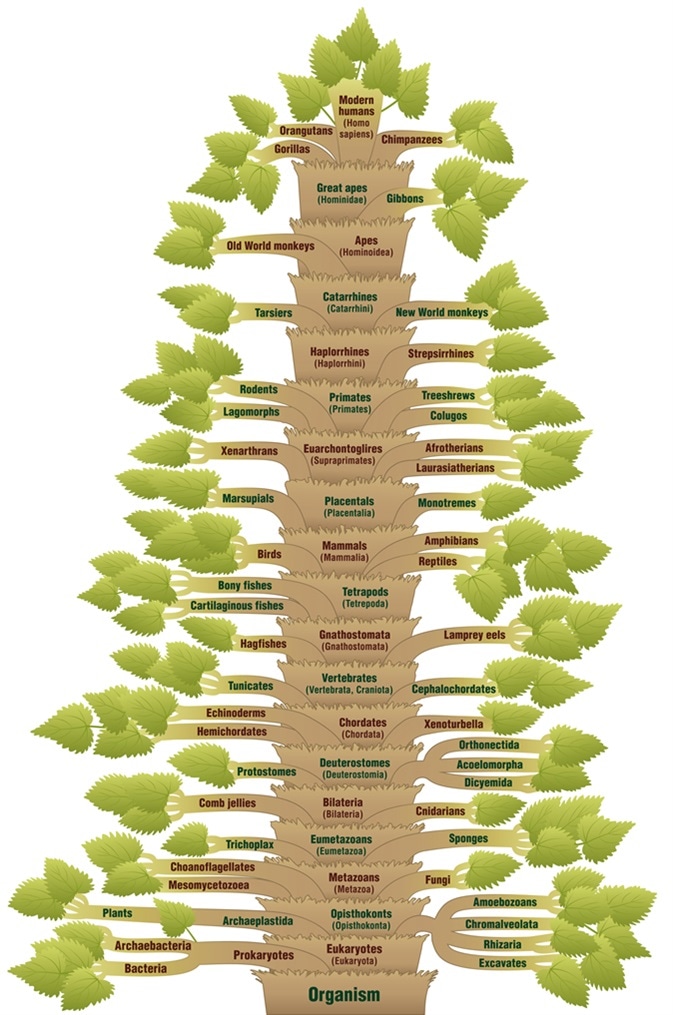
Detailed scientific classification of modern humans, from ORGANISM via VERTEBRATES through to HOMO SAPIENS. A phylogenetic tree with trunk (orders/suborders) and branches (related life forms). Image Credit: Peter Hermes Furian / Shutterstock
Taxonomy and Phylogeny
Taxonomy is the science of classification where biological organisms are grouped together and named based on shared characteristics. It allows for rational communication between scientists, including biologists and microbiologists. Phylogeny is a useful tool for taxonomists because it can be used to investigate evolutionary development. Taxonomy led to the study of phylogeny through the framework of dividing organisms into a hierarchy of taxonomic categories such as family, genus and species.
The classification scheme developed by Linnaeus in the 18th Century would later be used as a foundation for inferring phylogeny by interpreting the evolutionary relationships between taxonomic categories. Both taxonomy and phylogeny require the comparison of characteristics between organisms, with studies first utilizing morphological features and then progressing to molecular data.
Rooted and Unrooted Phylogenetic Trees
Phylogenetic trees can be rooted or unrooted. Rooted phylogenetic trees meet at a point upon a single node that represents a hypothetical common ancestor. There are various methods for producing a rooted phylogenetic tree but the most common utilize an outgroup consisting of a distant relative for all other species within the analysis. Rooted trees provide a representation of evolutionary relationships through time, with the longest pathways between a species and ancestor reflecting the greatest evolutionary distance between them. Unrooted trees display the connections between organisms without indicating ancestry and do not require a known or inferred ancestor.
Molecular Phylogenetics
Phylogenetic approaches require large datasets analyzed through rigorous mathematical modeling. Molecular data can produce a larger number of characteristics in comparison to morphological features. Nucleotides in DNA are unambiguous with character states of A, C, G and T that can be clearly defined. Morphological traits, in contrast, are based on shape and structure which can overlap and be difficult to distinguish. The simple conversion of molecular information to numerical form means that this type of data is particularly suitable for phylogenetic analysis.
A further complication of phylogenies produced through morphological data is phenotypic plasticity. This is where there is less constraint for a behavior or morphological characteristic to change in response to a unique environment. Phenotypic plasticity can therefore cloud phylogenetic signals. A number of surprising evolutionary relationships have been discovered through molecular phylogenetics, having not been highlighted by previous phylogenies produced by morphological traits.
Molecular phylogenetic trees are obtained by the comparison of nucleotide sequences. Homologous sequences indicate that they are derived from a common ancestral sequence. The DNA sequences are then aligned so the homologous nucleotides can be compared, noting any divergence by accumulation of indels and point mutations. The scored nucleotide differences are then used to reconstruct the phylogenetic tree with bootstrap analysis often performed to provide confidence limits.
Estimation of Speciation From Phylogenies
Phylogeny can also be used to estimate the duration of speciation, explaining how long it takes a new species to form and the factors influencing this time span. Simulations have found that phylogenies often do not contain enough information to produce unbiased predictions of speciation and extinction rates, but there are modeling methods that reduce bias. The protracted speciation model assumes that speciation is a gradual process with continual formation of incipient species experiencing a protracted pathway that can eventually lead to the development of a new species.
A single species is defined within the model as a complex of all lineages that have not yet been separated by a speciation-completion event. The protracted speciation model can explain reduced rates of lineage accumulation through time and the analysis of phylogenies, combined with the protracted speciation model, produces a novel approach to the estimation of speciation duration.
Sources:
- Brown TA. 2002. Genomes. 2nd edition. Oxford: Wiley-Liss; Chapter 16, Molecular Phylogenetics. https://www.ncbi.nlm.nih.gov/books/NBK21122/
- Money, N.P. 2016. 3rd edition. Academic Press; Chapter 1, Fungal Diversity. https://www.sciencedirect.com/science/article/pii/B9780123820341000013
- Soul, L.C. & Friedman, M. 2015. Taxonomy and Phylogeny Can Yield Comparable Results in Comparative Paleontological Analyses, Systematic Biology, 64, pp. 608-620. https://www.ncbi.nlm.nih.gov/pubmed/25805045
- Etienne, R.S. et al. 2014. Estimating the duration of speciation from phylogenies, Evolution, 68, pp. 2430-2440. https://www.ncbi.nlm.nih.gov/pubmed/24758256
Further Reading
Last Updated: Feb 26, 2019