May 18 2009
Researchers from Brown University and other institutions have developed a computational computer model of how brain tumors grow and evolve.
The model is the product mathematical formulas based on the first principals of physics, such as conservation of mass, and it has allowed researchers to recreate tumor growth in a computer. Through subsequent repetitive testing against real tumors, they have also linked their computerized tumors to real-world brain tumors, or "gliomas," and can now watch tumor growth on a computer screen.
Creating such a model is significant because it could help design specific, targeted treatments for individualized therapy. There is no cure for gliomas, which can kill quickly, often within 15 months of diagnosis. Massachusetts Sen. Ted Kennedy announced a year ago that he was suffering from this type of tumor, a malignant glioma of the brain.
Details of the research were highlighted at an April meeting of the American Association for Cancer Research. The full article is included in the May 15 edition of the journal Cancer Research.
"This helps us design a treatment," said Elaine Bearer, the lead author and professor of pathology and laboratory medicine at Brown. "By testing potential therapies in the computer, we can get our new drugs much faster to patients."
Bearer, who also has faculty appointments in the Division of Engineering and the Department of Music, worked with a number of collaborators on the project, including co-author and Vittorio Christini, a mathematician at the University of Texas.
To conduct the study, Bearer and her collaborators developed a mathematical formula that incorporated a number of equations describing the process of tumor evolution and growth. The master computational model was built on formulas that predict how much oxygen tumor cells consume and the rate of oxygen diffusion, and quantitative measures of cell growth and metabolic rates. The model is a series of interdependent differential equations. Each equation includes variables, or numerical values that can be experimentally manipulated.
For example, to test if oxygen consumption rates influence tumor growth, the values assigned to those rates can be changed and the outcome observed on the computer screen - not unlike playing a computer game. The result: A three-dimensional matrix of a glioma that can be adjusted to see what its growth stage will be over time, including speed of growth, size and shape.
Researchers validated their computational model with glioma specimens. Bearer studied about 40 different human brain tumor samples. Bryan Kinney, a member of Bearer's lab, obtained the samples from a number of sources, including the Rhode Island Hospital pathology department, the Columbia University Brain Bank and the Cooperative Human Tissue Network, a division of the National Cancer Institute that helps increase access to human cancer tissue for research.
The samples, sliced and sealed in a specimen slide, were as large as a chick pea or as small as the head of a pin. The tumor specimens used in the studies had been removed for diagnosis or surgical treatment of the tumor.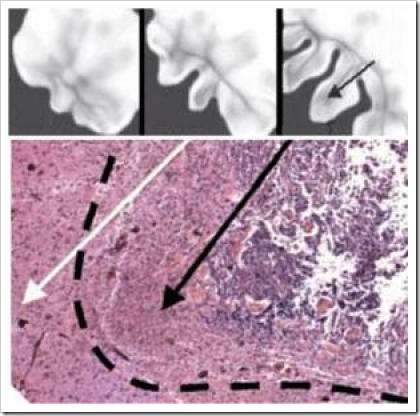
Researchers compared their virtual computational tumor with the actual human brain tumor samples at different stages of tumor evolution. Through many rounds of checking computer output against the real-life tumors, researchers created a computational model that mimicked natural biological tumors in all respects.
They focused specifically on a glioma because it does not invade the body through a basement membrane as epithelial-based cancers do, such as tumors that grow in the colon, breast or prostate. In the brain, tumors grow without having to digest a basement membrane to invade adjacent tissue. A basement membrane is essentially boundary of a given tissue that separates cells from the surrounding connective tissue.
Bearer said she hopes the research will allow doctors to find drug targets for glioma. She also envisions using the computational model to find targets for personalized medical therapies, enabling it to quickly identify molecular targets, and then select from existing treatments or design new treatments to stop the tumor.