Advancing the development of novel methods for understanding gene expression, Integrated DNA Technologies (IDT) has enabled Dr Jim Hughes, Associate Professor at the Weatherall Institute of Molecular Medicine, Oxford, UK, to optimize his unique Capture-C method, based on Chromosomal Conformational Capture (3C). As outlined in the recent DECODED Online article, Understanding How Distal Regulatory Elements Control Gene Expression, Dr Hughes uses IDT XGen® Lockdown® Probes to generate highly adaptable target capture probe pools for determining enhancer:promoter interactions, allowing greater flexibility in experimental design.
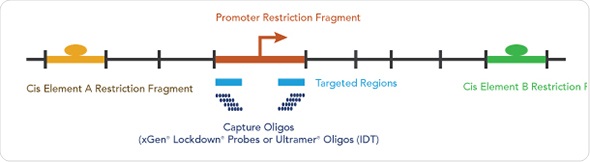
With enhancers and other regulatory elements often lying 10s to 100s of kilobases from the genes they affect, understanding the regulation of gene expression is an incredibly complex endeavor. The article describes how Dr Hughes and his collaborators have developed a versatile and scalable solution to enrich for enhancer:promoter interactions by integrating oligo capture and NGS technology into the 3C workflow. Off-the-shelf probe pools or panels often do not match a researcher’s experimental needs, while custom designed probe pools or panels often come at a high cost. Instead, Dr Hughes has utilized IDT’s xGen Lockdown Probes, scaled to create affordable custom probe pools to target specific regions of interest in the genome. He is able to modify the target and perform multiplex assays easily by mixing and augmenting pools with additional probes to create new panels, as required. Using this method, hundreds of genes can be interrogated in the same assay.
Developing this new method has allowed Dr Hughes and his team to enhance their workflow while saving costs as their research interests evolve. Their work has already brought new insight into the currently accepted “looping model” of promoter:enhancer interaction, and has secured Capture-C as a prominent method for addressing gene regulation.
Read the DECODED article to find out more about how Dr Hughes has used xGen Lockdown Probes to greatly improve his Capture-C protocol. For more information please visit www.idtdna.com/xgen. Follow us on twitter @idtdna for real-time updates and insights.
Source: Integrated DNA Technologies