RNA is becoming an interesting drug target as it takes possible intervention back one step to the synthesis of a target protein, instead of trying to block or inhibit a process.
Some work targeting RNA is attempting to stop RNA transcription, so that proteins cannot be translated, or to interrupt translation by blocking the recognition sequences in the RNA.
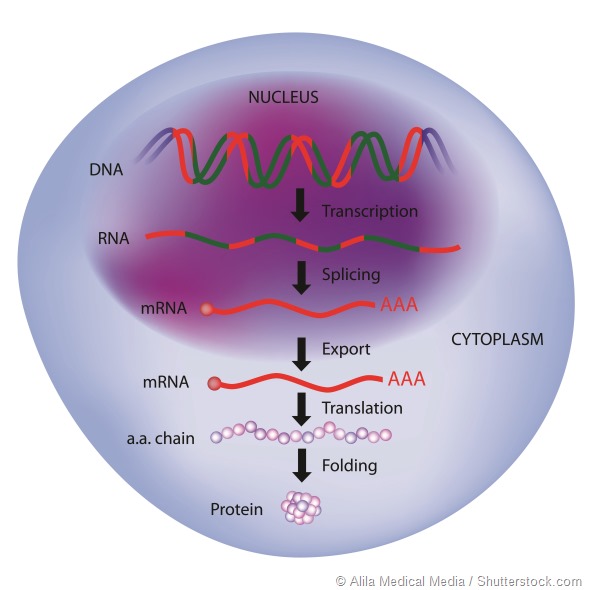
Both techniques are interesting, but may have problems in that complete absence of a target protein may result in compensatory mechanisms in the body that eventually overcome the effect.
Targeting alternative pre-RNA splicing is a different approach because it attempts to modify the RNA to change the protein that is made, rather than blocking it entirely.
In conditions such as spinal muscular atrophy, this means that a defective protein produced because a key exon is skipped could be replaced by block of that exon skipping.
Our approach is to try and change splicing of alternative exons, to change to function of the resulting protein.
What is alternative splicing?
There are many forms of alternative splicing, all of which can result in different RNAs and hence proteins generated from the same gene.
There is exon skipping, where an exon can be missed completely, intron retention, where intronic sequence is included, mutually exclusive exons, where, for example, either exon 2 or exon 3 are included in the RNA, but not both, or alternative 5’ (5 prime) or 3’ donor sites can be used, resulting in part of an exon being missed out.
It is the latter that interests us as the target family that we work on vascular endothelial growth factor, has two families that differ because one has exon 8a and the other exon 8b.
Do you think alternative splicing could provide a new approach to drug development?
It’s amazing how many proteins have variants that are generated by alternative splicing, and how many of those may dramatically change cellular function.
There are families of, for example, ion channels, where there may be hundreds of splice variants that could be profoundly affecting function of the brain.
So yes, once we understand more about the controls I think it will be a powerful new approach that could enable us to prevent changes caused by alternative spicing in diseases.
There are also many genetic conditions that result from aberrant alternative splicing – this is what has driven quite a lot of research in the clinical application in this area.
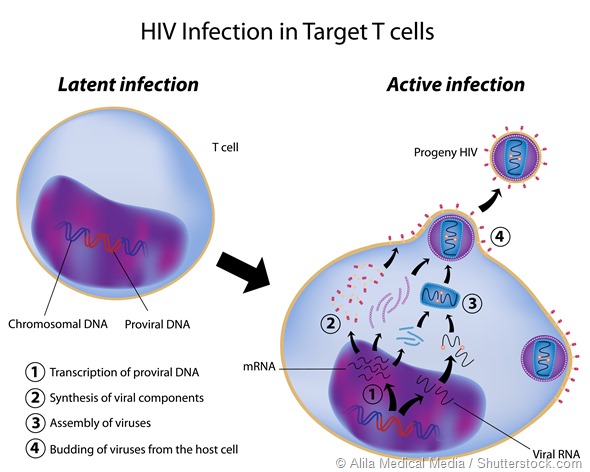
The other area that has driven a lot of work in alternative splicing is the study and control of retroviruses such as HIV. These pathogens depend on alternative splicing to generate the proteins used for replication etc., so there may be potential in that area as well.
What stage is the research currently at?
We have a lead candidate in our drug discovery programme for wet age-related macular degeneration, and we are currently in negotiations to raise investment so that we can take that into clinical trial. With the correct level of investment we aim to have that in clinical trial in 2 years.
The work in analgesia is behind the eye disease programme, and we are currently in the hit-to-lead stage in our drug development.
What that means is that we’ve demonstrated that we can change the alternative splicing of the pre-mRNA we are targeting in appropriate laboratory tests, including in cells, and that when we do that we can affect pain in laboratory animals.
Now we are optimising our novel drugs to ensure that we have one that can have these effects on pain, with the right degree of potency, without any side-effects, and in a form that is easy and convenient to take.
How many clinical trials are currently targeting alternative splicing? What can be done to increase this?
There are several currently in clinical trial. There are two clinical trials for treatment of Duchenne’s muscular dystrophy, and three in spinal muscular atrophy (target SMN2). These are both conditions where exons are known to be skipped, resulting in the clinical effects, and splicing inhibition is being targeted to try and correct that.
Unfortunately, although the therapy is still being pursued in Duchenne’s the Phase 2 trial failed to meet its outcomes. The trials for SMA have also run into some difficulties including some toxicity issues, but splicing control is still being pursued in SMA as well.
There are also clinical trials in myeloma, cholangiocarcinoma, cystic fibrosis (Target CFTR), diabetes (insulin receptor) and familial dysautonomia (exon skipping in the kinase complex associated protein gene), but results are not yet out for these.
What are the main challenges that will need to be overcome?
At the moment, although we understand some of the ways in which some alternative splicing is controlled, there is a lot we don’t know about how cells ‘decide’ that a pre-mRNA will be spliced in one way or another. We really need to get a better understanding of those processes, and particularly those that are specific to a disease, or condition.
If we target ubiquitous mechanisms then we run the risk of getting in the way of normal function, which could be disastrous. There’s a lot to be done in this area, as the specific controls are only really well understood for a very small number of targets.
On a personal level, we need to overcome the challenges of funding the work on the pain programme. Funding is really tight all over the world, and that is particularly true for translational work such as this.
Experimentally, getting the right mode of action for a new drug, doing the work to show that it works and that it could be clinically useful is tough.
We have an excellent team of scientists across the world, including in Australia, so we have the right people working on this, and we have the backing of the University, our spin-out company Exonate Ltd and our investors. This means we are well positioned to push the research forward, but we still have a lot to do.
What do you think the future holds for alternative splicing as an approach to drug development?
It is an entirely new approach to altering function that has not been tried in drug development for human RNA splicing, so it is very exciting.
It has to be approached with care; as I said, if drugs affect constitutive splicing, that is the normal splicing together of exons that occurs in the vast majority of mammalian mRNAs then that would be disastrous, as it would halt normal cellular functions.
If we have the understanding of the specific ways in which important disease-causing alternative splicing events come about, then it could give us very specific and effective treatments.
Where can readers find more information?
Our first publication on alternative splicing control and pain can be accessed here: http://www.sciencedirect.com/science/article/pii/S0969996114002435
and work from our collaborators on alternative splicing in eye disease:
and cancer:
Information on our spin-out company, in partnership with the University of Nottingham is here:
About Lucy Donaldson
- Associate Professor, University of Nottingham (2013 -)
- Senior Lecturer, University of Bristol (2003-2013)
- Lecturer, University of Bristol (1999-2003)
- Lecturer, University of Leicester (1996 – 1998)
- Postdoctoral research fellow, University of California at Davis (1994-1996)
- Doctoral research fellow, Pharmacology and Medicine, Faculty of Medicine, University of Edinburgh (1990-1994)
- Bachelor of Dental Surgery (1985-1990)
- Bachelor of Neuroscience (1988-1989)