Please can you give a brief introduction to head and neck cancers?
Head and neck cancers (HNC) are the sixth most common cancers worldwide, with approximately 600,000 new cases diagnosed every year.
One of the challenges in HNC studies relates to the variety of anatomical subsites that comprise this disease, which include tumors of the nasal cavity, paranasal sinuses, oral cavity, nasopharynx, oropharynx, hypopharynx, and larynx.
Despite recent advances in therapeutic approaches, the overall 5-year survival rate of HNC patients has not improved over the past decades.
The major known risk factors associated with these carcinomas are cigarette-smoking, alcohol consumption, and in some cases, the presence of etiological agents, such as HPV and EBV.
Which type of cancer does your research focus on and how much is currently known about the molecular basis for this cancer?
In our laboratory we currently have a couple of HNC proteomics projects. In particular, my interest and research over the past few years has been focusing on cancer of the oral cavity, specifically tongue squamous cell carcinoma (TSCC).
TSCC are characterized by aggressive behavior, with a high number of patient developing recurrence, lymph and distant metastasis. The molecular mechanisms that drive tumor progression and invasion in TSCC are not well understood, it is, therefore, of great importance to understand which pathways drive carcinogenesis and disease progression, in order to develop novel therapeutic interventions for TSCC.
My research interests over the past few years have been focusing on the identification of signaling pathways within the tumor microenvironment that support TSCC cancer progression.
By using an in depth MS-based proteomic approach, we have been able to identify proteins secreted from cancer-associated fibroblast (CAFs) that can act as paracrine regulators of TSCC growth.
In this experimental model, the proteomics analyses of fibroblast secretome have been integrated with plasma membrane proteomes of TSCC cells.
Our goals are to provide insights into the functional role of a pro-invasive stroma and a mechanistic link between complex microenvironmental stimuli and phenotypic tumor response.
Why is it important to understand the activation of fibroblasts?
Overall there is a great interest in understanding the differences between normal stroma and reactive tumor stroma, mainly because the active stroma provides oncogenic signals that can promote tumorigenesis.
Fibroblasts are one of the main components of the stroma and the principal source of matrix deposition. What we know is that reactive stroma has been associated with an increased number of fibroblasts and there are studies indicating that fibroblasts can affect cancer progression by secreting proteins, as well as extracellular vesicles, which carry oncogenic molecules able to affect cancer cell behavior.
Please can you outline some of the hypotheses for the origin of the cancer associated fibroblasts (CAFs)? Why is this still controversial?
One of the main challenges in the study of cancer associated fibroblasts is the lack of accurate definition of their phenotype/functional markers, together with the limitations associated with the experimental models which are unable to reflect the same level of complexity as seen within the tumor microenvironment in vivo.
CAFs are extremely heterogeneous populations of cells that harbor different characteristics in relation to 1) the tissue type, 2) the paracrine factors involved in CAFs activation and 3) the cell type of origin. For these numerous reasons the origins of CAFs is still controversial and might not be easily answered.
One important source of CAFs are resident quiescent fibroblasts that evolve together with the growing tumor and acquire a modified phenotype (CAF-like). The definition of the CAF-like state has been associated with some characteristics such as, increased α-SMA and FAP expression, ECM deposition and metalloproteinase activity, but none of them seems to be specific enough and further research is needed to define more accurate markers or a panel of molecules associated with the CAF-phenotype.
Other possible sources of CAFs can be found in cancer cells that undergo EMT and acquire mesenchymal properties, or tumor activated bone-marrow cells that have entered the tumor site.
How is the tumor microenvironment defined and why is it so important?
The tumor microenvironment can be described as a complex and multidirectional network of interactions between the different cell types as well as non-cellular components surrounding the tumor. It consists of fibroblasts, activated fibroblasts, immune cells, endothelial cells, components of the extracellular matrix, soluble factors, pH, etc. Taken together, all these factors have a major role in influencing the outcome of the malignancy.
Bidirectional communication between cells and their microenvironment is crucial for both normal tissue homeostasis and for tumor growth. Numerous studies have shown how such cross-talk is mediated either by cell-cell interactions or soluble factors, and recently it has been highlighted how important the role of other key players such as miRNAs, metabolites and exosomes are.
What are exosomes and what role do they play in cancer?
Exosomes are nanometer-sized extracellular vesicles that range between 40-200 nm in size, they are secreted into the extracellular space and can horizontally transfer biologically active molecules (proteins, lipids, and nucleic acids) to recipient cancer cells (or vice versa).
Tumor-derived exosomes can modulate several functions in the surrounding stromal cells (fibroblasts, endothelial cells, immune cells) and promote their activation, angiogenesis and metastatic processes. On the other hand, CAF-derived exosomes are also reported to play significant roles in promoting and regulating cancer progression.
Therefore, a better understanding of CAF-derived exosomal protein content together with the investigation of their functional effects on tongue cancer cells can give novel insights on the complex molecular interactions underlying stromal-tumor crosstalk and help to elucidate their roles in regulating carcinogenesis.
Please can you outline your research on isolating CAFs and proteomic analyses of CAF and AF secretome and exosomes?
To investigate CAFs characteristics in TSCC we have isolated matched pairs of human primary fibroblasts from resected tumors (CAFs) and adjacent tissue (AFs) and characterized them according to established CAF markers.
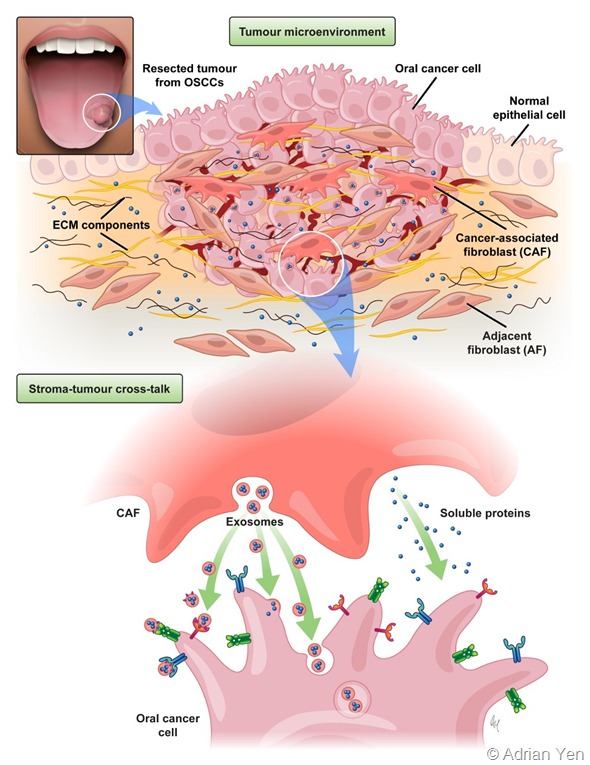
Image credit: Adrien Yen
We looked at differences in their morphology, α-SMA expression and the activity to degrade extracellular matrix components. Based on these criteria we have screened 9 matched pairs of CAFs and AFs that we have used for our proteomics analyses as well as for functional validation experiments.
We then have employed a shotgun proteomics approach to identify CAF-associated proteins in three different fractions, total lysate, conditioned media and exosomes. The comprehensive proteomics dataset we have generated, consists of 4247 proteins, has allowed us to identify signatures of CAF-enriched proteins potentially involved in pathways mediating tumor-stromal crosstalk.
How did you evaluate the effects of fibroblast-derived exosomes on tongue cancer cells?
In addition to our proteomics finding, to assess the functional relevance of exosome-mediated crosstalk, we have shown a significant effect of CAF-derived exosomes on TSCC cells motility, proliferation and the activation of the MAPK pathway. Of note, this effect was proven to be completely ablated following heat denaturation suggesting that the biologically active cargo is a protein(s).
Can you please describe how you have generated a signaling map of fibroblast-cancer cell ligand-receptor interactions? What impact will this have?
To deepen into the fibroblast-tumor cells signaling, we have combined our fibroblast secretome data (conditioned media) with an in-depth repertoire of plasma membrane proteins that we have successfully generated from TSCC cell lines.
We have isolated and characterized TSCC surface proteome by using a protocol that has been optimized in our lab, and utilizes colloidal silica-beads coupled with MS-based proteomics.
We are currently working on a network map we have generated as result of these global analyses. This map represents a useful ligand-receptor repertoire that will allow us (and others in the field) to validate these interactions and to identify specific oncogenic pathways and molecular targets in the oral cancer microenvironment.
Where can readers find more information?
For more information about Kislinger Proteomics Lab please visit http://kislingerlab.uhnres.utoronto.ca/
Readers can also access the Webinar on “Proteome Profiling of the Tumor Microenvironment: Role of Human Primary Fibroblasts Derived Exosomes in Oral Cancer Progression” at the following link https://www.labroots.com/ms/webinar/proteome-profiling-tumor-microenvironment-role-human
About Dr Simona Principe
After receiving her Master in Medical Biotechnology (2007) and PhD in Immunopharmacology (2011) from the University of Palermo (Italy), Simona Principe is currently conducting postdoctoral research in the lab of Dr. Thomas Kislinger at the Princess Margaret Cancer Center in Toronto, Canada.
Her research interest focuses on applying proteomics technologies to investigate cancer biology. In particular, she studies the role of secreted extracellular vesicles as novel intracellular signaling mediators within the tumor microenvironment that support cancer progression and metastasis. Since 2012 she has published 5 papers/reviews in the field of proteomics and exosomes.