The term dark proteome refers to proteins whose structural features and thus functions are not well understood.
Many proteins within the dark proteome do not fold into stable three-dimensional structures. These proteins are called intrinsically disordered proteins (IDPs) and feature highly flexible, disordered confirmations.
The structures of IDPs cannot be represented as static molecular images and therefore are largely unseen, constituting the “dark” portion of the proteome.
Why do certain proteins not adopt defined 3D structures?
Intrinsically disordered proteins exhibit amino acid sequences that don't allow them to collapse into unique three-dimensional structures.
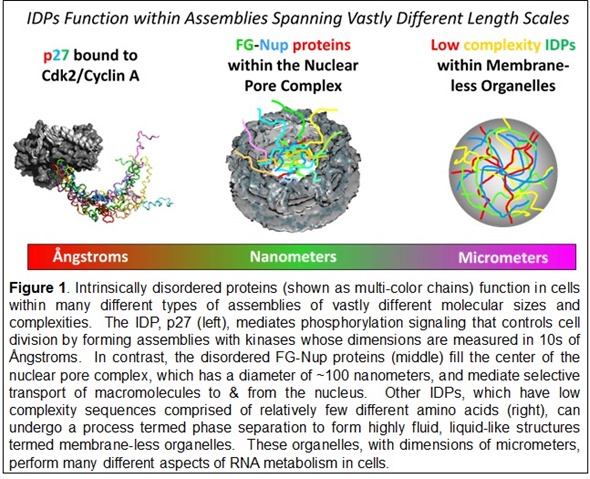
© Richard Kriwacki, Ph.D. and Darcie Miller, Ph.D. of St. Jude Children’s Research Hospital
Such proteins are enriched in amino acids not typically found in the cores of folded proteins—hydrophilic amino acids as well as negatively and positively charged amino acids.
The sequences of some IDPs exhibit repetitive patterns of just a few amino acids termed low complexity sequences. These features are associated a phenomenon termed phase separation. The process underlies the formation in cells of micron scale, liquid-like structures termed membrane-less organelles.
The features of the IDPs that form the liquid-like structures radically depart from the traditional paradigm of protein structure, illustrating how IDPs have diversified the repertoire of protein-containing structures in cells.
How many of these proteins exist and what are their functions within cells?
Humans have about 20,000 proteins. Thirty to 50% are believed to be either entirely disordered or to have disordered regions.
We're able to make these estimates because, as previously noted, disordered proteins have amino acid compositions that are different from folded proteins.
IDPs play amazingly diverse roles in cells, ranging from signaling and regulation in cellular communication networks, to mediating contraction in muscle, to serving as “glue” that holds membrane-less organelles together.
Does that mean that they have a lot of different functions?
Yes, they have what I describe as myriad function. It's almost limitless.
You can start from the nucleus of the cell – many transcription factors that regulate gene expression are disordered proteins. And there are disordered regions within the histone proteins that make up nucleosomes, the structural unit of chromatin.
These disordered regions are called histone tails. They mediate the epigenetic code because they have residues that can be modified in different ways, including phosphorylation and methylation, and control how the information in our genome is read by the gene transcription machinery. This is another important example of disordered proteins.
There are proteins in the nuclear pore complex, which is a gateway that mediates the movement of biomolecules into and out of the nucleus. The diffusion barrier, or gateway, in the center of the nuclear pore is comprised of disordered proteins that form a liquid-like mesh that allows only certain proteins and other biomolecules to pass through.
In the cytoplasm, there are all sorts of regulatory disordered proteins that interact with membrane receptors.
As mentioned above, disordered regions of proteins also play a role in formation of membrane-less organelles through a process called phase separation. When these proteins achieve a certain concentration in a cell they self-associate. The result is a dynamic meshwork that constitutes the "structure" of the membrane-less organelles.
Researchers recently showed that under certain circumstances the IDPs within some membrane-less organelles can convert to rigid, fibril-like structures that are toxic to cells and are associated with certain neurodegenerative diseases such as amyotrophic lateral sclerosis, or ALS. These observations highlight the importance of IDPs in not only in human biology but also human disease.
Why has there been limited research into these “intrinsically disordered proteins” (IDPs) so far?
I think there are two fundamental reasons why the research community hasn't given IDPs as much attention as they deserve.
In the mid-90s there was a bias in the structural biology community against the idea that proteins could perform functions by being disordered.
There are still structural biologists who will acknowledge that while proteins in isolation may be disordered, such proteins actually become rigid and adopt discrete structures when they perform their cellular functions. While this is true for some IDPs, it is not for many, many others.
In several examples of biological functions or settings, where disorder is mediating important processes, the IDPs remain dynamic and disordered as they function.
The FG-Nup proteins within the nuclear pore complex are a striking example. These proteins fill the center of the pore and form a dynamic, mesh-like barrier that allows some but not all biological macromolecules to pass from the cytoplasm to the nucleus, or vice versa. The flexibility and disorder of the FG-Nups are essential for their gating function within the pore.
The other fundamental reason that IDPs are understudied is that characterizing their features is difficult—they are constantly moving!
New, integrative approaches involving traditional and non-traditional structural biology methods are being applied to relate the dynamic features of IDPs to their functions. However, right now few labs are prepared to undertake such studies.
But the tide is turning, and more and more labs are entering the field. It is extremely important, though, that government and other funding agencies invest funding in this exciting area to accelerate progress.
How have recent developments in technologies such as nuclear magnetic resonance (NMR) spectroscopy impacted our understanding of IDPs?
NMR is particularly powerful because it allows, at the amino acid level and the individual atom level of resolution, the observation of signals for both folded regions and disordered regions of proteins. It really has been a major driver, especially in defining the features of disordered proteins in isolation.
However, NMR can also be used to study disordered regions of proteins with large molecular assemblies—for example, with liquid-like droplets that reflect the way IDPs are organized within fluid membrane-less organelles.
What do you hope to learn about IDPs?
The major goal of the Dark Proteome Initiative that we launched recently is to draw attention to the importance of disordered proteins and attract funding.
The funding would support basic science to discover the fundamental functional mechanisms of disordered proteins through multidisciplinary investigation to understand their structural features and how these disordered structural features are related to function. This will allow for much broader descriptions of how proteins perform their biological functions.
Right now, for a large portion of the proteome that we describe as being dark, including dark proteins, we simply don't understand the relationship between their physical features and their functions.
A major goal of the Dark Proteome Initiative is to eliminate this knowledge gap and provide information that will establish how proteins function in cells. Importantly, that would help us understand what goes wrong with these proteins in various human diseases.
What diseases are known to be affected by IDPs?
I think the one association that's quite well understood is that with neurodegenerative diseases. It is well-established that degenerative diseases such as Alzheimer's disease, Parkinson's disease, amyotrophic lateral sclerosis (ALS), and Huntington's disease arise from the aggregation of disordered proteins into small oligomers that are toxic to cells.
There are many ways in which disordered proteins are associated with cancer. One example is the transcriptional regulatory protein Myc. Myc is an IDP that partially folds upon binding another protein, Max, and DNA.
Myc is over-expressed in a wide variety of cancers. Cancer cells select for over-expression of Myc in order to take over the transcriptional machinery of the cell to drive proliferation. That's one type of mechanism by which a disordered protein is associated with cancer.
Yet another mechanism relates to the fact that many cancers arise through gene fusion events that bring together one portion of a gene for one protein with a portion of the gene for another protein. The resulting fusion proteins have aberrant functions. In many cases, one of the partners in the fusion protein is an IDP. This occurs in a wide variety of leukemias, sarcomas and various other cancers.
Another association with disease is in the arena of infectious diseases. Many viruses express disordered proteins that take over the regulatory machinery of cells. These viral regulatory proteins actually draw upon the endogenous regulatory features of naturally disordered proteins found in the host cells but optimize these features for pathogenesis through rapid cycles of viral evolution.
What do you think the future holds for unlocking the dark proteome?
I think we are on the brink of a new era in the fields of structural biology and cell biology. In the next decade, we will gain deep, mechanistic understanding of how this important portion of the proteome functions.
Gaining this fundamental knowledge is going to provide a basis for understanding a wide variety of diseases. That knowledge will lay the foundation for discovery of innovative therapeutic strategies to combat disordered proteins in a wide variety of diseases.
Structure-based drug discovery is a major contributor to the development of new therapeutics. To target a protein (e.g., an enzyme associated with a disease), you define a nice binding pocket, then find a small molecule that can fit into that pocket, and inhibit the enzyme or inhibit its interaction with other proteins. But disordered proteins require a different approach.
There are examples of small molecules that modulate the function of disordered proteins. In several cases, small drug molecules block the binding of an IDP to a folded protein partner and therefore inhibit the IDPs function. Some of these molecules are right now in or moving toward clinical trials.
Another strategy the pharmaceutical industry has taken is to use therapeutic antibodies against the pathological oligomers (made of IDPs) that are associated with the development of neurodegenerative diseases. However, the results of these efforts have been mixed but progress has recently been made.
In the future one could also imagine antibodies developed to target specific parts of disordered proteins with regulatory features that have gone awry in different diseases.
Another possible strategy is to use synthetic genes as therapeutic agents. It would involve introducing synthetic genes that encode disordered proteins, possibly synthetic disordered proteins with novel, beneficial functions. The synthetic genes and artificially constructed disordered proteins could be used to combat the disruption of disordered protein function associated with certain diseases.
Where is the field heading? There are three essential elements—discovering how these proteins work; understanding the connection between IDPs and disease; and then leveraging the knowledge to develop innovative therapies to combat diseases involving disordered proteins.
Where can readers find more information?
- Dunker AK, Kriwacki RW. The orderly chaos of proteins. Sci Am 304(4):68-73, 2011. PMID: 21495485. Translated for German and French affiliates of Scientific American.
- Mitrea DM, Kriwacki RW. Phase separation in biology; functional organization of a higher order. Cell Commun Signal. 2016 Jan 5;14(1):1. doi: 10.1186/s12964-015-0125-7. PubMed PMID: 26727894; PubMed Central PMCID: PMC4700675.
- Babu MM, Kriwacki RW, Pappu RV. Structural biology. Versatility from protein disorder. Science. 337(6101):1460-1, 2012. PubMed PMID: 22997313.
- Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signaling and regulation. Nat Rev Mol Cell Biol. 2015 Jan;16(1):18-29. doi: 10.1038/nrm3920. Review. PubMed PMID: 25531225; PubMed Central PMCID: PMC4405151.
About Richard W. Kriwacki, PhD
Member, Department of Structural Biology
St. Jude Children's Research Hospital
Memphis, Tennessee USA
Richard Kriwacki, Ph.D., is a member of the St. Jude Department of Structural Biology. Dr. Kriwacki’s research focuses on understanding the functional mechanisms of proteins involved in regulation of cell division, apoptosis and ribosome biogenesis and how these regulatory mechanisms are altered in cancer and other catastrophic diseases. Many of the proteins involved in regulating these critical cellular processes are IDPs, which has fueled Dr. Kriwacki’s longstanding interest in this area of protein research.
His recent publications include research published in eLife detailing the role of a protein named Nucleophosmin in the liquid-like structure of a membrane-less organelle called the nucleolus. The molecular machines that synthesize proteins, termed ribosomes, are assembled in the nucleolus and the recent article described how Nucleophosmin helps to bring the many different components of ribosomes together within the liquid-like nucleolus for assembly. Another recent publication in Scientific Reports detailed evidence that small molecules bind and inhibit the disordered protein p27Kip1. As a disordered cell cycle regulatory protein, p27 was not previously considered to be a viable drug target. Dr. Kriwacki has published widely in other peer-reviewed journals including Cell, Science, Molecular Cell, Nature Chemical Biology, Nature Structural & Molecular Biology, and Proceedings of the National Academy of Sciences.
Kriwacki serves as an adjunct professor in the Department of Microbiology, Immunology and Biochemistry at the University of Tennessee Health Science Center. He earned a doctorate in chemistry and biophysics from Yale University, a Master of Science degree in Pharmaceutical Sciences from the University of Connecticut, and performed postdoctoral research at the Scripps Research Institute in La Jolla, California. He is currently a member of the editorial board of the Journal of Molecular Biology.