Alzheimer's disease (AD) diagnosis and treatment has been a challenge and this is mainly due to the varying degrees of degeneration and cognitive damage seen in different patients. The researchers from the University of California San Diego School of Medicine for the first time used the transcriptome to assess the proteins made from the genes in patients with AD, that could help in diagnosis.
A transcriptome is the sum of all the messenger RNA (mRNA) molecules that are expressed by the genetic code of an individual. The study is published in the latest issue of the journal Cell Reports, this week and is titled, “Integrating gene and protein expression reveals perturbed functional networks in Alzheimer’s disease”.
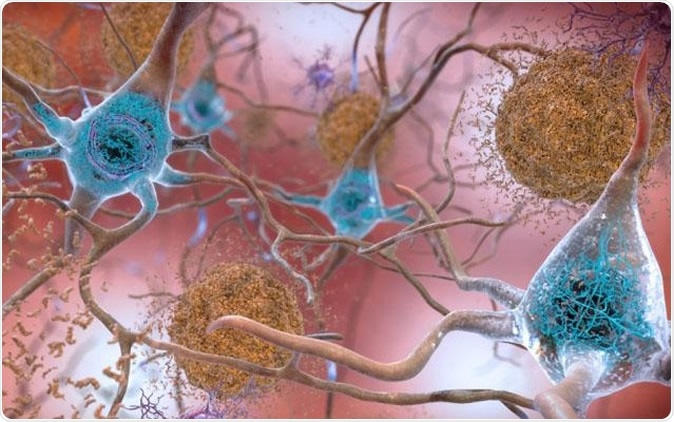
In brains affected by Alzheimer's disease, abnormal levels of the beta-amyloid protein clump together to form plaques (seen in brown) that collect between neurons and disrupt cell function. Abnormal collections of the tau protein accumulate and form tangles (seen in blue) within neurons, harming synaptic communication between nerve cells. Image Credit: National Institute on Aging, NIH
For their study the team of researchers compared the transcriptome from 414 study participants. These persons had been diagnosed earlier clinically with Alzheimer's. Their genetic make-up was compared with an age matched control participant who was of the same age but did not have features of dementia. Results revealed that there are a complex interplay between genetic alterations and the interplay of various proteins in AD and this could help in diagnosis of this condition even when features of cognitive decline and neurodegeneration is absent.
Results revealed that those with a family history of AD tend to have a strong genetic connection with the disease which was not surprising. On the other hand those with sporadic AD have multiple factors that determine their condition. Many of these factors are unknown. Sporadic AD remains the more common form of AD say the researchers. Some of the risk factors include gender, age and other social, biological and psychosocial factors.
The team explains that there is accumulation of two types of protein in the brain and this is the key pathology of AD. These proteins are formation of amyloid protein plaques and development of abnormal neurofibrillary tau protein tangles. These plaques and tangles tend to kill the neurons and lead to the features of AD and its progression. Other factors such as inflammation, brain atrophy and blood circulation problems have all been attributed to AD and its progression, the authors of the study add. The team wrote that these factors, “correlate with clinical symptoms of cognitive decline and have led to changes in diagnostic criteria during the last decade.”
The team wrote that some of the genetic mutations linked to AD that are aready known include, “Genetic mutations in amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2) associated with autosomal dominant AD.” The team added in their study that loss of the regulation of “EGR3” gene could be responsible for the synaptic deficits or the deficiencies in the nerve synapses in AD. They explain that this could be due to targeting the synaptic vesicle cycle which is altered in AD. They proposed, “loss of regulation of EGR3, which is crucial for short-term memory, mediates synaptic deficits by targeting the synaptic vesicle cycle,” and its association with AD in their study.
Senior author Robert Rissman, PhD, professor of neurosciences at UC San Diego School of Medicine, explained that their study looked specifically at genetic alterations and related protein interactions in AD. Rissman is also the Director of the Biomarker Core for the Alzheimer's Disease Cooperative Study (ADCS) and of the Neuropathology/Brain Bank and Biomarker Cores for the UC San Diego Shiley-Marcos Alzheimer's Disease Research Center.
He explained that these genes are composite clusters that are identifiable and their level of expression is related with AD. He added that these genes could be affecting the cell cycle, metabolic processes within the cell, immune responses and ultimately cell survival. These factors are all connected with the pathology of AD he explained. The team wrote that these clusters of genes were identified using “Louvain algorithm for community detection”.
The authors of the study explained, “The Louvain algorithm does not require selection or fine-tuning of parameters, as do some other clustering algorithms such as, K-means clustering and hierarchical clustering (such as are used in the WGCNA R package). Rather, clusters are determined by finding groupings of genes which have many within group connections, and few between group connections. This algorithm has proven useful in detecting modules, or clusters, in protein-protein interaction networks.”
Rissman in a statement said, “One of the big problems in AD research is identifying patients at risk at the right time. Understanding the gene networks that may change in specific patient groups can help streamline clinical trials recruitment efforts and reduce costs and time to enroll trials. With the field shifting more and more toward pre-symptomatic disease, we need to expand our understanding of the molecular mechanisms that underlie the entire disease spectrum.
Journal reference:
Integrating Gene and Protein Expression Reveals Perturbed Functional Networks in Alzheimer’s Disease
Canchi, Saranya et al. Cell Reports, Volume 28, Issue 4, 1103 - 1116.e4, https://doi.org/10.1016/j.celrep.2019.06.073