Researchers from the Massachusetts Institute of Technology (MIT) have made a scientific breakthrough where their machine-learning algorithm or Artificial Intelligence (AI) program successfully created a novel antibiotic compound. The drug has since then been tested on deadly bacteria in the lab, and in lab mice with a considerable amount of success, explain the researchers. The study results appear in the latest issue of the journal Cell.
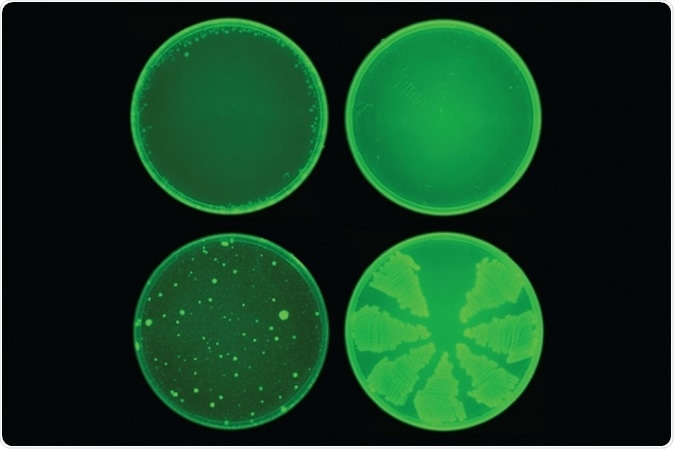
MIT researchers used a machine-learning algorithm to identify a drug called halicin that kills many strains of bacteria. Halicin (top row) prevented the development of antibiotic resistance in E. coli, while ciprofloxacin (bottom row) did not. Image: courtesy of the Collins Lab at MIT
Antimicrobial resistance is a menace gripping the world with a large number of strains of bacteria developing resistance against the currently used antibiotics, making simple infections life-threatening. Search has been on to develop new antibiotic compounds to kill these resistant strains of bacteria.
The MIT researchers created a computer program that could screen through a hundred million potential chemicals that could be antibiotics and choose the appropriate molecule. This molecule would have the properties necessary for it to kill the bacterial strains. The mechanism by which these novel molecules kill the bacteria is entirely different from the currently available antibiotics, making them a surer weapon against the infections, added the researchers.
Lead researcher James Collins, the Termeer Professor of Medical Engineering and Science in MIT's Institute for Medical Engineering and Science (IMES) and Department of Biological Engineering, said, "We wanted to develop a platform that would allow us to harness the power of artificial intelligence to usher in a new age of antibiotic drug discovery. Our approach revealed this amazing molecule which is arguably one of the more powerful antibiotics that has been discovered."
Researcher on the team, Regina Barzilay, the Delta Electronics Professor of Electrical Engineering and Computer Science in MIT's Computer Science and Artificial Intelligence Laboratory (CSAIL), said, "The machine learning model can explore, in silico, large chemical spaces that can be prohibitively expensive for traditional experimental approaches." This meant that this approach could help researchers screen molecules for their antibiotic potential without wasting resources on failures of lab tests with molecules that may not work. This study was authored by Jonathan Stokes, a post-doctorate student at MIT and the Broad Institute of MIT and Harvard.
What was done?
For this new study, the team looked at several possible molecules that the algorithm could screen. They then zeroed in on the potential molecules with suitable properties using AI. The model, they wrote, could be used to design and learn the chemical structures of the drugs to make new antibiotics. After finding the likely candidate molecule, the team tested it in the lab in the in vitro studies. They then replicated their findings in lab mice infected with strains of bacteria. In both experiments, the candidate molecule showed significant success.
For their experiment, the team first taught the computer program the possible molecules which could be deadly to a common bacterial strain of E. coli. The program scanned through 2,500 molecules of which around 1,700 molecules were FDA approved, and 800 were naturally derived products. Once trained, the program was used to scan through 6,000 compounds in a library created by the Broad Institute's Drug Repurposing Hub.
From these 6,000 compounds, the computer program found one molecule which had a unique chemical structure but could be lethal to the bacterial strains. The team of researchers called the molecule "halicin." Halicin was named after the AI system from "2001: A Space Odyssey". This molecule has been investigated before as a drug for the treatment of diabetes.

The original prop of the HAL 9000 from the Stanley Kubrick adaptation of 2001 A Space Odyssey. It is on display along with other pieces at the Design Museum in Kensington. Image Credit: Hethers / Shutterstock
Halicin was found to be effective in killing lab-grown, and patient-derived colonies of bacteria such as Clostridium difficile, Acinetobacter baumannii, and Mycobacterium tuberculosis which were difficult to treat using conventional antibiotics. They noted that bacteria Pseudomonas aeruginosa was not responsive to halicin.
Next, the team infected lab mice with Acinetobacter baumannii that was usually resistant to all known antibiotics. They used halicin ointment on the mice and found the infection to be cleared within 24 hours. They explored the mechanism of the drug action and noted that it could act via altering the electrochemical gradients across the cell membranes of the bacteria. This mechanism has not been studied before and thus could prevent the bacteria from developing resistance, explained the researchers.
Way forward
Collins explained that there is a growing need for new antibiotic molecules that work differently in order to fight antibiotic resistance. He said, "We're facing a growing crisis around antibiotic resistance, and this situation is being generated by both an increasing number of pathogens becoming resistant to existing antibiotics, and an anemic pipeline in the biotech and pharmaceutical industries for new antibiotics." He explained that this approach had been tried before but without success. This was the first time that an actual drug could be developed and tested in the labs with reasonable success.
Stokes added, "When you're dealing with a molecule that likely associates with membrane components, a cell can't necessarily acquire a single mutation or a couple of mutations to change the chemistry of the outer membrane. Mutations like that tend to be far more complex to acquire evolutionarily."
The team plans on working on Halicin and other candidate molecules with a pharmaceutical company or nonprofit organization, in order to commence clinical trials and use them safely and effectively in humans in the near future.
Journal reference:
A Deep Learning Approach to Antibiotic Discovery Stokes, Jonathan M. et al. Cell, Volume 180, Issue 4, 688 - 702.e13, https://www.cell.com/cell/fulltext/S0092-8674(20)30102-1