Many drugs consist of small molecules. In order to find new drugs, researchers need to find molecules that bind to cell targets such as proteins. In the current study, they utilized the power of VR to understand how the molecule acts on proteins, by replicating the world inside the protein for the program user. This experience helps to identify the atomic-level interaction between the protein and the small molecule drug. The type of VR used is called interactive molecular dynamics simulations in VR (iMD-VR).
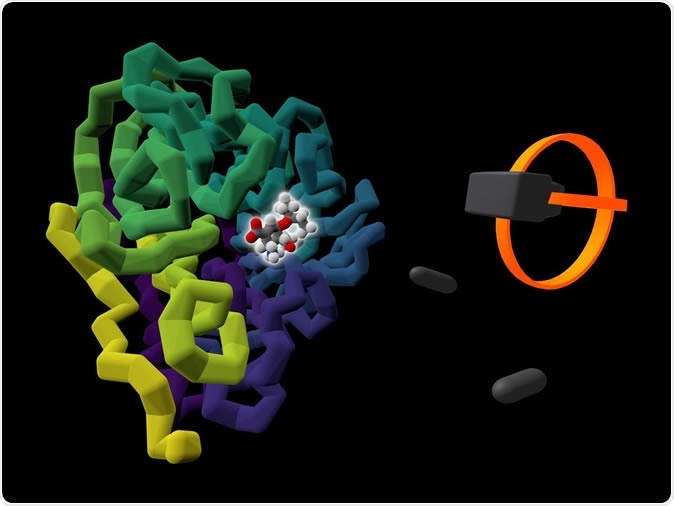
The commercially available influenza drug Tamiflu (highlighted by a glow) is shown bound to the viral protein it targets, neuraminidase (colored in purple to yellow). A rendering of a user's VR headset and controllers is presented alongside the protein. Image Credit: University of Bristol
The study
The researchers used the iMD-VR strategy to simulate the attachment of a drug molecule to a protein, to be able to predict in advance exactly how the two would be attached. They looked at several drugs, including those used for flu and HIV.
The binding of a drug to a protein is crucial in inhibiting the function of the protein by more than one possible mechanism. One instance is when a virus is prevented from replicating itself within the host cell by a drug that binds to one of the viral proteins.
For firm binding, such a drug, composed of a small molecule, must have a snug fit. The challenge before the researchers is to identify such molecules that are capable of this type of tight binding. This can help to design improved drugs.
The contribution of VR to this process is explained by investigator Adrian Mulholland, "Researchers need to understand how drug molecules fit into their biological targets. To do this, we use VR to represent them as fully three-dimensional objects. Users can then fit a drug within the 'keyhole' of a protein binding site to discover how they fit together."
Binding the drug, predicting the structure
The team first set the VR users to the task of binding drugs to viral protein targets, such as the neuraminidase of the influenza virus and the protease enzyme of HIV. By simulating the three-dimensional structure of the protein, the VR users were able to accurately predict the binding of each of the given drugs to their protein targets. In addition, they came up with drug complex structures that closely resemble the experimental structures, by simply maneuvering the drug into the protein.
The most impressive thing was that laypeople could pull in the drugs to attach correctly to the simulated protein using VR. The conclusion is that interactive VR can help tell how a new molecule with potential therapeutic effect will perform concerning binding its target protein.
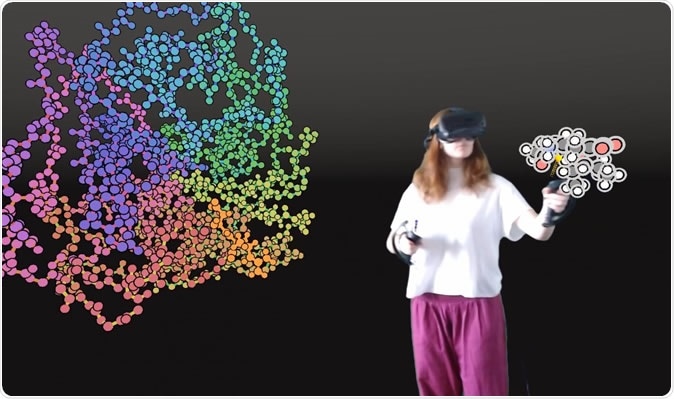
User interacting with a protein in VR. Image Credit: University of Bristol
Benefits of the VR approach
The VR strategy used in the study uses commonplace equipment and an open-source platform, making it easily available for any user, lay, or expert.
Thus structure-based drug design is now possible for anyone interested in the process, using VR. In the words of Mulholland, "An important aspect of the work is that the drugs, and their protein targets, are fully flexible: we model their structural changes and dynamics, and users can manipulate them interactively to find how drugs interact with their biological targets. This is a really exciting and powerful way to model drug binding. We have shown in this work that it gives accurate results."
Other researchers agree, pointing out that simulation allows drug-protein binding and repeatedly unbinding, with a timeframe that is markedly less compared to the time required for similar phenomena when the users use non-interactive molecular dynamics VR. Expert David Glowacki says, "The full unbinding and rebinding events generated using iMD-VR were achieved by the users in less than five minutes of real-time."
Moreover, the use of VR allowed non-expert users to bind, unbind, and then achieve a docking position that closely resembled the initial structure, sufficient to consider that they had re-docked the drug. All that was needed for this was the provision of trace atoms which showed the right pose for the drug.
If such trace atoms were absent, Mulholland remarks: "Binding poses understandably had more variation, but users were still able to get within the same range of the accepted bound position for all three systems.:
And to cap it all, he says, "These results were achieved within a single hour-long training session with each participant, demonstrating the usability of this VR framework." This shows the immense potential to apply this technology to drug design in an extremely cost-effective, rapid, and highly productive manner.
Journal reference:
Interactive molecular dynamics in virtual reality for accurate flexible protein-ligand docking Deeks HM, Walters RK, Hare SR, O’Connor MB, Mulholland AJ, et al. (2020) Interactive molecular dynamics in virtual reality for accurate flexible protein-ligand docking. PLOS ONE 15(3): e0228461. https://doi.org/10.1371/journal.pone.0228461