We have been working in the domain of the visceral emotional neuraxis for some time. This visceral emotional neuraxis is where there is an interplay between the state of peripheral organs, mood and mental function, and so on.
It is well known that stress has a detrimental effect, particularly at the heart and probably at other organs. There is a whole system of communication between the brain and the organs that subserves this and allows this interplay between visceral state and emotional state.
The health of the peripheral organs depends on that interplay operating properly. When it does not, you get, for example, heart failure and heart disease. At the same time, it has been demonstrated under some conditions that you can improve a diseased organ by improving the way that the innervation of the organ is operating by getting it into a more healthy domain.
These are all interesting scientific and clinical questions, but it is a topic that has not gotten a huge amount of support and funding. It tends to, as they say, fall between two stools. It is not quite neurology, and it is not quite cardiology. It is there and it is known about, but it is not really focused on.
A program came along called SPARC. It is not focused on neurology or the heart, or lungs, blood or any specific traditional area. It is an overarching scheme that tries to address opportunities that are not being chased by specialist organizations.
A large program was put together with the goal of providing a comprehensive mapping of the relationship between autonomic outflow and, particularly, the vagus nerve and peripheral organs, including the heart, which we are interested in.
The initial phases of it were foundational. The goal was, "Let's see what this is all about. How is this thing organized? How does it work?", and to use those answers as a springboard for getting into more clinical studies. We applied and eventually were able to receive some support, and this work is the byproduct of the contribution to that effort.
It has been a fairly longstanding interest to get to the function beyond the structure. This first attempt looked at the anatomical and molecular structure of the system. Ultimately, the goal is to figure out the function of the system.
Why are neural processes within the heart such an important aspect of bodily function? And why was it so important to map this neural system?
Heart disease is very common and by far the biggest cause of death worldwide. People suffer mightily from heart disease. They may have a considerable amount of pain for some time or be very reduced in their functionalities.
The pharmaceutical approaches that have been taken so far have been somewhat efficacious. They are not worthless, but they are far from addressing the problem.
The development of devices to manipulate the activity of the peripheral nerves would potentially be a great positive influence in understanding and dealing with organ health - in this case, heart health.
It is really important to unpack the neural computation and circuit part surrounding organs in order to be able to better view, design, and test these devices and make them safe. So far, we do not know enough to be able to make those devices effective and safe.
The key to this is to develop a detailed anatomical molecular and functional map of the system of the heart itself. For example with the vagus nerve, you could essentially stop the heart completely if you altered this in the wrong way.
What is the intracardiac nervous system and how does it function?
In one route, the central nervous sends an axon out to its target and innervates the muscle or the skin directly. That trafficking and communication is direct and back and forth between the peripheral target and the central nervous system.
However, what often surprises people is that there is a parallel nervous system called the autonomic nervous system in which that is not the case. In the autonomic nervous system, there are neurons interposed between the nerves coming out from the brain and the end organ.
So in our case of the heart, that means that there is a whole nervous system that is intermediate, like a little brain in the heart. It is the effector that is affecting the heart, not the brain.
This is something that is not actually that widely known, even within cardiologists. The neurons tend to be embedded in fat pads on the atria and in the ventricles in humans.
All the positive effects that the outflow from the brain has on the end-organ must pass through these intermediate neurons embedded in the heart.
People have been studying these neurons, but there has been far less than a comprehensive appreciation of their number, organization, distribution, and certainly not their molecular identities and phenotypes. We are beginning to fill in these gaps.
Raj carried out research that showed that there is a tremendous amount of local activity within this system, that does not depend on the brain. The heart does communicate sensory stuff information with the brain, but on a moment to moment basis, it has a closed-loop system taking care of things locally.
We looked for control properties and showed that there is much potential for local computation in a way that simplifies the problem for the brain to regulate. We were not dealing with a comprehensive map like we are today.
Now that we have this complexity, we want to see if we can turn that into a dynamic understanding of how all these molecules lead to certain controlling properties. What kind of local computation happens? There are some fascinating questions to chase, now that we have this comprehensive foundational substrate.
In the next phase, one of the things that we are doing is working with Peter Hunter in Auckland, New Zealand, who is developing an abstraction of the heart which he called a scaffold.
We are mounting our mapping into that scaffold in such a way that there is now an independent objective 3D representation of the heart and of the heart's nervous system, in such a way that other data can be integrated into it.
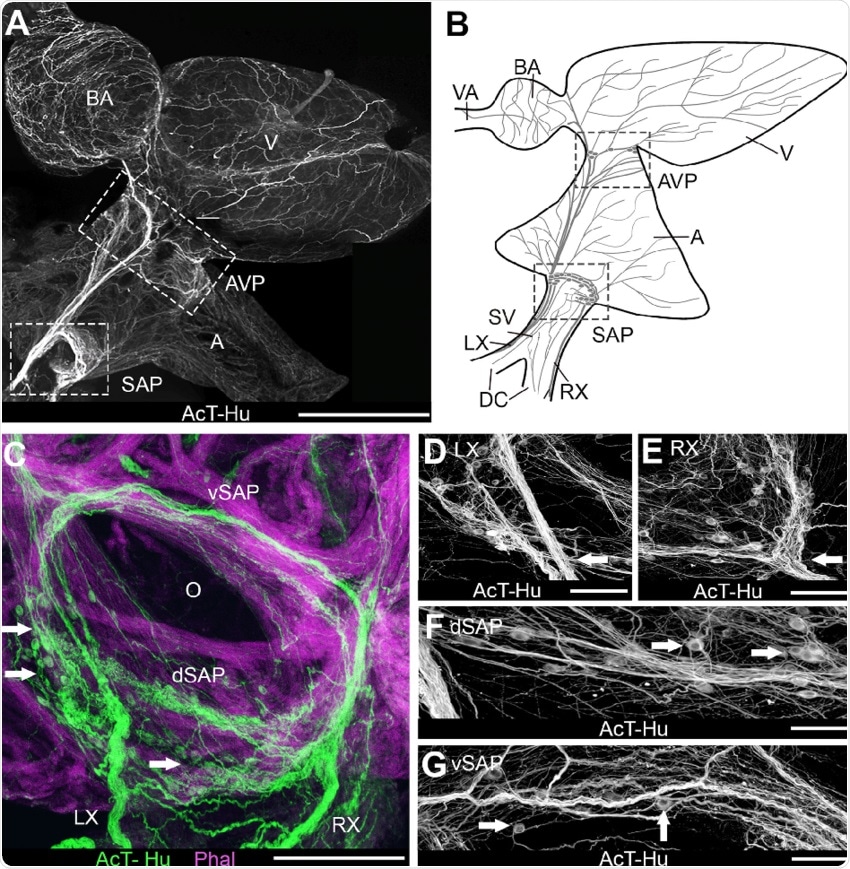 Image Credit: Intrinsic and extrinsic innervation of the heart in zebrafish ( D anio rerio ): Zebrafish Cardiac Innervation - Scientific Figure on ResearchGate. Available from: https://www.researchgate.net/figure/Organization-of-intracardiac-nervous-system-demonstrated-with-acetylated-tubulin-AcT_fig1_272844021
Image Credit: Intrinsic and extrinsic innervation of the heart in zebrafish ( D anio rerio ): Zebrafish Cardiac Innervation - Scientific Figure on ResearchGate. Available from: https://www.researchgate.net/figure/Organization-of-intracardiac-nervous-system-demonstrated-with-acetylated-tubulin-AcT_fig1_272844021
What is knife-edge scanning microscopy (KESM) and how did you use it?
A small company in South San Francisco called 3Scan developed this KESM technology.
We had just taken a rat heart and taken something called the cryostat, a device that takes a larger piece of tissue like a heart and turns it into tissue sections that can be mounted onto glass slides for looking at under the microscope. In doing this we were a little bit limited by the fact that the heart has got these giant chambers that can collapse in different ways.
What we wanted to achieve was to take the data that is present at each slide, as stained neurons, and stack those sections along with the mapping of the neurons back into a 3D representation of the whole organ with the correct positioning of those neurons. But when each one of those sections is distorted to a fair extent, the outcome is not accurate.
One of the things that cause this problem is called the banana problem. If you have a banana and it has got a curve, and you cut it into thin slices and then put the slices back together by stacking them you are going to lose the curve – they are just going to be straight up and down. It is not going to look like a banana.
What knife-edge does is take a diamond knife that is transparent meaning that the microscope can see the tissue through it. The diamond knife is sectioning at five microns, which is pretty thin. A microscope camera system is then capturing digital images of the section as it is being sectioned, at a resolution of about a half a micron by a half a micron pixel size.
Due to the very high-resolution image of the tissue section in situ, you are capturing a detailed image of the section itself which means that the stack is going to be exactly the same as the original organ with no distortions. That is knife-edge scanning microscopy.
The second part of the problem is that there is a lot of data. If you are doing it manually with whole slice imaging, depending on how the slide is imaged even the stacking may not be that straightforward to do and the alignment becomes a problem even after you have a reference frame to get around the banana problem.
What the KESM system was doing was not only acquiring images within one reference frame but setting the whole system up for high data throughput. The mass of data that you collect and sort is integrated into the whole KESM system in such a way to make a downstream analysis much easier. That makes a world of a difference.
How did you build your 3D model of a rodent heart?
KESM was the first part of it, acquiring the rat hearts and getting the image data. That was quite a large volume of image data.
We have a second collaborator called MicroBrightField, MBF Biosciences. Their specialty is software for 3D mapping of tissue sections from the brain and subsequent 3D reconstructions of regions of the brain.
However, they modified their software to work with the heart. Using their TissueMaker™ and Tissue Mapper™ software, they were able to take data from 3Scan and turn it into a stack of images mappable by the software.
We have other collaborators in Orlando, Florida at UCF, as well as ours in Philadelphia, who had used that software to create these mappings and these 3D representations.
It has very much been a joint effort. A lot of teamwork has been involved, for which I am proud of. The key was that everybody was sold on what the goal and purpose of this is. To get to that 3D map at a very high resolution was a unique once in a lifetime opportunity. An essential element was to let the data speak for itself and not bias it with what we thought the biology ought to be.
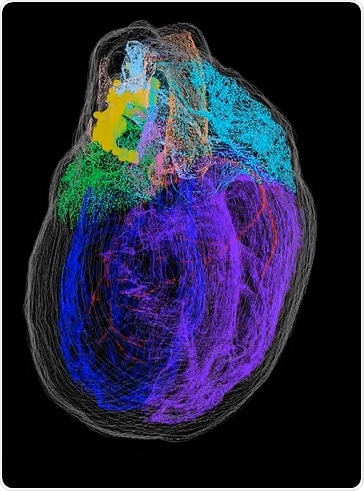
What did you find in terms of neuronal distribution in relation to anatomical structures of the heart? How have you applied this to the functions of these anatomical structures?
The rat heart is the most widely used model of physiology, pharmacology, and biochemistry. It has similarities to humans but is also widely different.
It is important to have a clear picture of the organization of neurons in the rat. The prior literature tends to focus on what is called the base of the heart. It seems counterintuitive, but the base of the heart is the top of the heart. This comes from embryology as the heart grows from the top down, and so the base is the origin of the heart, where it grows from. Then it grows down to the so-called apex at the bottom of the ventricles.
The base of the heart (hilum) is where the focus of this neural system has been. The hilum of any organ is the region in which the vessels that serve it enter and exit. So you have arteries and veins – the pulmonary artery, the aorta, pulmonary veins, and so on going in and out of this region.
In development, the neurons migrate into the heart, down the vessels, and then just scatter out. The thought was that you are going to wind up with this very variable, disorganized, and haphazardous scattering of neurons.
This is not what we actually saw when we were able to put the thing back together. There is quite a lot of innervation coming from the hilum around the vessels, especially with the pulmonary veins and around the vena cava.
These clusters are almost like a continuous sheet of neurons that extend out of the hilum. One of the places they go to is the thin membrane separating the atria called the interatrial septum.
The clusters continue down the left atrium on the posterior surface of the heart, and those clusters are substantial. They are not continuous - they are distinct clusters that extend all the way to the ventricular boundary. That was a big surprise.
The next thing that we have done looked across quite a few different hearts. What we found is that they are actually quite consistent with one another and it is not a random or wildly variable situation at all.
Having a full map starts opening up new possibilities to say: Are these clusters the same? How are they different? In what ways can they be exhibiting distinct functions, or are they there for redundancy? Suddenly the way we ask those questions might change because the path at least has been refined in a very substantive way.
What did you find out through gene expression analysis of individual neurons?
One of the important questions before going into this was what is the level of complexity of these neurons? Are they mostly based on one or two types of molecules that they use to communicate? Are they cholinergic? Do they use adrenaline or adrenergic neurons? Most of the literature that has been out there describe it as a two-part system.
For a long time, these things were thought of as a nuance that could not do much, because the brain must be where everything happens.
Here, we went into single neuron gene expression, and that in itself was a technically challenging feat. Laser capture microdissection was needed and most importantly spatially tracking where we were getting these neurons from so that they could be put back together into the map.
When looking at that gene expression that is spatially tracked, there is a tremendous amount of complexity. There is a wide range of neurotransmitters in the neuromodulatory system present. And so, the potential for computation is immense. On top of it, there were not distinct types of neurons that they broke down into distinct neuropeptides. There were not distinct sets of neurons that made distinct neuromodulators. We see a combinatorial logic at play.
The way we have to think about the problem is not that there are completely distinct parts that somehow come together, but rather every neuron has some capability to do many, many things. Multiple neurotransmitters and neuromodulator processes can be activated and flexibly used in each neuron.
And so, how would that system operate? It opens up a lot of new questions into how we think about plasticity and adaption within this process. There is a story that connects this to the brain and molecular gradients.
We are following behind the technologies that have been recently developed in the brain. The leader there was the Allen Institute.
They have developed technologies together with MicroBrightField (MBF) and have been the leader in figuring out how to make these kinds of molecular atlases. What they have found consistently is that the neurochemical properties or the molecular phenotypes are present in gradients throughout the different parts of the cerebral cortex. You see modulators and transmitters and receptors distributed in gradients.
That is what we were looking for at the heart as well. And indeed, there are gradients. It is not like all the neurons do the same thing. The distribution of cardiac neurons are differentially expressing modulators and transmitters and receptors.
They do so in an interesting way that suggests that in addition to direct neuron-to-neuron function, there could also be opportunities for paracrine function, that is, a cluster of neurons might be bathed in a modulator so as to alter the population activity. This provided opportunities to start thinking about interventions much more broadly.
It was weakly known that there was more at the heart than acetylcholine and norepinephrine, but fundamentally that is how people have thought about the system – that the neurotransmitter that is used by the neurons at the heart is acetylcholine and there is a modulation from norepinephrine inputs, but that is about it.
Through our research, the heart neuronal system has been shown to be more interesting, more complex, and more organized than previously thought. There looks to be a fundamental structural organization to the heart's nervous system both in terms of its clustering, its distribution, and the kinds of modulatory factors and receptors that it employs.
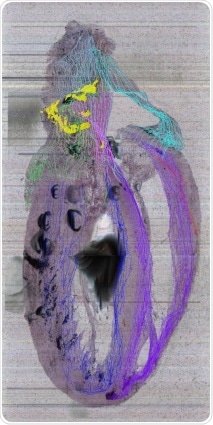
Up until now, the only other organ with such a detailed neuronal 3D map is the brain. How could the protocol you have used help to detail other organs, and how could this help improve scientific and medical knowledge?
MBF Biosciences was our exclusive collaborator and subcontractor on our grant for doing the heart. However, once the SPARC leadership saw what we were getting, they snatched them off into what they call MAPcore, where they are working with all the other organs in the SPARC program, such as the pancreas, the bladder, and the intestine.
Also, there were so many lessons learned in working with the heart, not just in KESM and with MBF Biosciences, but with how to annotate and share that information and turn it into a more generalizable scaffold.
The vision for SPARC is to have all this data in a single, highly structured, annotated, and accessible resource that is available to the community in every way. The hope is that this will be a heavily used resource that will get populated with additional relevant data sets that have used it.
How far away do you think 3D neural imaging of a human heart is?
The only thing that stands between us and that is money.
There are a few technology scaleups that have to be made, but none of them are insurmountable. It is very clear what it is that we have to do, and we have given it much thought. A lot of detailed planning has occurred on how one might go about that.
We have even scaled some of this to a small chunk of a pig heart. The pig heart is huge - it is as big as a human heart and it is more similarly organized to a human heart. It is not exactly the same, but it is a good scaleup.
We think the human heart will be done in less than a decade, given that so many things have been figured out in the pipeline and the key is timing. As with anything, when people gather around, put their collective brains and technologies together to work on a problem and it is all fresh in the mind, that is the time to do it.
Image Credit: Africa Studio/Shutterstock.com
How could this model and subsequent human models influence our understanding of cardiac health and disease?
One is the therapeutic or protective effects of the vagal activity on the heart passing through these neurons. If you could associate the protective or therapeutic activity to specific neurons and modulators, you could emulate that either by finding the right stimulation parameters or feeding with the right molecular manipulations.
Another step that would really help augment our efforts to develop interventions is getting a sense of dynamics of the system. We need to develop into how these parts move about and interact with each to yield certain network dynamics.
When we develop that initial understanding, that would help prioritize and narrow down the intervention possibilities instead of just relying on static anatomical snapshot data. This is a substrate on which dynamic simulations and models will be built.
What are the next steps for your research?
We have explored the variability across hearts in both males and females, a paper that is just about done. In another direction, we have begun to explore mounting these data sets into Peter Hunter's Auckland Bioengineering Institute scaffold.
We have also started to build dynamic models of the system to examine what control properties they exhibit, and how they would play in circuits together with the central nervous system. That is the grant we do not have funding for but hope will be funded July 1st.
I think this SPARC effort out of the common fund in the Director's Office has been successful and is doing what it intended to do, which is to begin to make not a third stool between the two, but a bridge.
Hopefully, we are heading in a direction where the individual institutes will then invest in making people cross those bridges and connect two disparate fields towards improving human health in ways that have not been possible before this.
Where can readers find more information?
Read the iScience paper here: https://doi.org/10.1016/j.isci.2020.101140
SPARC portal: https://sparc.science/
About Dr. Vadigepalli
Rajanikanth Vadigepalli, Ph.D., is Vice-Chair of Research and Professor of Pathology, Anatomy, and Cell Biology at Thomas Jefferson University.
Dr. Vadigepalli’s collaborative research program at the Daniel Baugh Institute for Functional Genomics/Computational Biology is driven by a convergence of systems engineering, computational modeling, bioinformatics, and single-cell scale transcriptomics, to identify and target key control points for intervention in disease.
Ongoing international collaborative projects focus on central and peripheral neural circuits controlling the heart, brainstem neuroimmune processes leading to hypertension, liver regeneration in alcoholic liver disease, and cell fate regulation underlying developmental defects.
About Dr. Schwaber
James Schwaber, Ph.D., is Director of the Daniel Baugh Institute for Functional Genomics and Computational Biology and Professor of Pathology, Anatomy & Cell Biology.
Dr. Schwaber uses systems biology approaches in the mammalian brain to study adaptive neuronal processes.
His main interest is in the emotional–visceral neuraxis and disorders involving this interaction, including those related to stress and autonomic imbalance in neurogenic contributions to hypertension, addiction and withdrawal from the dependent state, and neurodegenerative conditions including epilepsy.