In this interview, News-Medical had the opportunity to speak with Dr. Xiaobing Han, Principal Scientist for Antibody Discovery at Rapid Novor. The discussion revolved around the significant potential of polyclonal antibody sequencing and other technologies in advancing the field of antibody discovery.
The field of antibody discovery and generation has undergone significant technological advancements, leading to the rapid growth of antibody therapeutics in the field of human and animal medicine. What are some of the latest technological advancements for antibody discovery?
The ability to isolate and sequence individual B cells is a more detailed and specific analysis of B cell diversity. Characterization of single B cells provides a direct link between the sequences and cells, allowing for the discovery of antibodies with specific properties, such as high affinity to a target, potent neutralization of pathogens, or involvement in autoimmune responses.
Library design for phage display and other display technologies have improved structural and sequence diversity, allowing for the development of antibodies against hard-to-target antigens. Libraries derived from immunized donors can yield antibodies with improved expression levels, affinity, and specificity for the target antigen, compared to naive libraries. On the other hand, the generation of synthetic libraries allows for complete control over antibody sequences through de novo design. This, in turn, can help mitigate and eliminate unfavorable sequences, ultimately saving time in downstream engineering and optimization processes. Recent advancements in library design involve sequence-based optimization aimed at improving the developability properties of antibodies, such as stability and solubility.
Image Credit: ShutterStock/ustas7777777
In silico modeling is becoming more broadly used as a method for screening, selecting, and or optimizing prospective antibody sequences after a discovery campaign. This approach enables the rational design of antibody characteristics, such as binding sites and binding kinetics. It can also be used to predict biophysical interactions, antibody aggregation, and stability. As such, in silico modeling may reduce the reliance on expensive trial-and-error experimentation.
Despite these advancements, several challenges persist in the process of antibody discovery today. Could you please elaborate on some of the main challenges currently faced in antibody discovery?
Hard-to-target antigens and novel drug targets are a major challenge in antibody and drug discovery. Targets such as transmembrane protein receptors account for 20-30% of the human proteome and make up 60% of current drug targets. Generating functional antibodies against transmembrane receptors is difficult due to their multi-domain complexity, conformational variability, low immunogenicity, and lack of soluble forms.
When utilizing in vivo models for antibody discovery, the suitability of animal models poses problems for early development and downstream testing. Immune tolerance, especially if the host expresses the target antigen in its own tissue, can result in challenges with eliciting an effective immune response. Cross-reactivity is particularly important to consider in pre-clinical and safety testing. Antibodies developed in animal models can cross-react with similar antigens in unintended ways due to similarities in protein structures, sequence homology, or expression patterns between species.
Similarly, immunogenicity, especially when producing antibodies within an animal model intended for human use, poses major safety and efficacy concerns. Differences in post-translational modifications, antibody structure, and amino acid sequences can incite an immune response against the therapeutic itself, creating anti-drug antibodies and can potentially trigger adverse reactions.
Image Credit: ShutterStock/Corona Borealis Studio
Accessing a diverse and functionally relevant antibody repertoire is a significant technological challenge in antibody discovery. Take B-cell sequencing, for example. While 2-3% of B cells in the peripheral blood originate from the bone marrow and spleen, this represents only a fraction of the overall B cell population. Only a minor proportion of these circulating B cells differentiate into antibody-secreting plasma cells. Consequently, relying solely on B cell sequencing to explore the antibody repertoire imposes limitations on the ability to tap into the entire antibody repertoire, encompassing upwards of 1012 clones. To visualize this, consider a handful of sand (10,000 grains) compared to all sand in a sandbox - that’s an approximation of the size of a B-cell sequencing campaign vs the whole antibody repertoire.
Ideally, during antibody discovery campaigns, it is beneficial to obtain a diverse selection of antibodies with variable functionality that target diverse epitopes. While an antibody discovery attempt can yield binders to the target antigen, it may be challenging to discover antibodies that have the desired functionality, such as agonist or receptor-blocking antibodies.
Identifying antibodies from discovery campaigns is only a portion of the pipeline. While selected antibodies display the ideal functionality, specificity, and affinity, they may not be suitable candidates in terms of developability. Degradation, aggregation in solution or upon administration, as well as formulation, poses issues for the successful development of an antibody candidate. Many campaigns are challenged with cost-effective characterization, developability analysis, and prediction in the discovery stages for successful antibody therapeutic development.
What strategies or approaches can be employed to effectively mitigate risks associated with antibody discovery, considering cost and speed?
Rigorous target validation is crucial in order to focus resources on the most promising therapeutic areas. Prioritizing well-validated targets with strong biological rationale increases the likelihood of success and reduces the risk of investing into targets with limited therapeutic potential.
There is no ‘one-size fits all’ approach to antibody discovery. Strategies that have proven effective in other discovery attempts may not yield the same successful results again. Adopting a multi-faceted strategy, with multiple immunization approaches applied across various species in tandem, increases the likelihood of success. Additionally, employing multiple discovery modalities concurrently, such as polyclonal antibody sequencing with B cell sequencing and in silico modeling, can increase the chances of success. Not only does this combine the advantages of these methods, but it also can accelerate the early-discovery phase of therapeutic development. In silico analysis and AI-driven antibody discovery in parallel with in vivo methods, it becomes feasible to anticipate and prioritize potential antibody candidates. This approach may reduce the need for extensive experimental testing and streamline the discovery process.
Image Credit: ShutterStock/Kateryna kon
Utilizing polyclonal antibody sequencing with REpAb technology provides an antibody discovery avenue that can accelerate the discovery process. Following immunization, the discovery process immediately starts off with highly abundant and functionally significant antibodies collected in serum. Utilizing a proteomics-based approach allows for antibody sequencing, followed by recombinant expression, to perform characterization and biophysical analyses. This approach negates the need for cell sorting, immortalization, and purification prior to characterization.
Antibody characterization proves most effective when conducted at regular intervals throughout the antibody discovery and development pipeline. Employing techniques to assess both biophysical and functional characteristics, as well as cross-reactivity and performance in the early stages of discovery, ensures that costs are not lost when candidates fail during cell line development and formulation.
High-throughput antibody screening assays are valuable for rapidly evaluating a large number of potential antibodies, enabling the selection of candidates with the desired properties. Utilizing high-throughput methods for kinetic analysis with surface plasmon resonance (SPR) allows for rapid analysis of a range of kinetics and affinity. As such, selecting a range of affinities, on-rates, and off-rates, as opposed to selecting the tightest binders, may expand the pool of antibody candidates. In the same regard, performing epitope binning and epitope mapping experiments with hydrogen-deuterium exchange mass spectrometry (HDX-MS) allows access to information on a diverse binding epitopes for downstream selection. Finally, early cell line development efforts are often described as a ‘needle in a haystack’ search and should be augmented with rapid analysis of sequence production and PTMs such as glycan profiles to improve success rates - real-time peptide mapping and glycan profiling by MS is an ideal tool here.
By combining these strategies, researchers can streamline and improve the efficiency of antibody discovery campaigns, maximizing the chances of identifying successful therapeutic candidates and setting up early production for success.

Image Credit: Rapid Novor Inc
What impact does AI and machine learning have in antibody discovery?
These technologies already play a crucial role in enhancing various aspects of antibody discovery.
In terms of target identification, AI-driven algorithms can analyze vast amounts of biological data, such as proteomics, genomics, and disease pathways, to identify potential therapeutic targets.
The generative AI buzz has created excitement for de novo antibody design, entirely generated by algorithms, without experimental data or relying on biology. However, this approach has yet to be proven as effective due to limited training data, the complexity of the antibody structures and functions, as well as the biological context and variability of the immune system.
In your recent breakthrough, Rapid Novor has been able to sequence human polyclonal antibodies directly from serum using only proteomics. Can you tell us how the mass spectrometry-based antibody discovery platform, REpAb, works?
Yes, this was a big win for Rapid Novor. Leveraging our antibody discovery platform with polyclonal sequencing, we successfully sequenced and expressed twelve full-length antibodies directly from the serum of a patient who had received the COVID-19 vaccine. The antibodies expressed through recombinant methods exhibited a strong affinity for the antigen and demonstrated neutralizing capabilities. Our polyclonal sequencing platform for antibody discovery proved its effectiveness in extracting protein sequences from a small human serum sample, enabling the discovery and production of functionally relevant antibodies.
Our antibody discovery pipeline begins with creating an immunization protocol tailored to the target host species. REpAb is a species agnostic antibody discovery platform; we have worked with humans, sheep, dogs, alpaca, goats, chickens, and rabbits and have the flexibility to adapt to more.
Following immunization and plasma collection, we perform affinity purification with chromatography to purify the antigen-specific polyclonal mixture from the plasma. Through the use of carefully selected proteases, the complex pAb mixture undergoes digestion into short peptides, generating hundreds of overlapping sequences. We conduct numerous liquid chromatography with tandem mass spectrometry (LC-MS/MS) experiments and other MS techniques to extract valuable information from regions of significance, such as complementarity-determining regions (CDRs).
With the data obtained, we employ AI-based bioinformatic algorithms to de novo sequence and assemble short peptides into longer peptides, ultimately generating full antibody heavy and light chains, including paired heavy and light chain combinations. The reported results contain full-length antibody sequences, as well as paired heavy and light chains, in addition to the CDR cluster sequences.
All of this can be done using a proteomics-only approach, negating the need for genomic information. However, we also have the capabilities to combine transcriptomics data captured from peripheral blood mononuclear cells (PBMCs) to further broaden the antibody discovery search. This enables us to further broaden the scope and depth of our antibody discovery efforts.
We conclude our REpAb discovery programs by recombinantly expressing the antibody sequences we’ve obtained and subsequently characterizing them. We screen and profile antibody leads with SPR for binding kinetics and HDX-MS for epitope mapping. We may also perform functional assays such as cell-based neutralization assays.
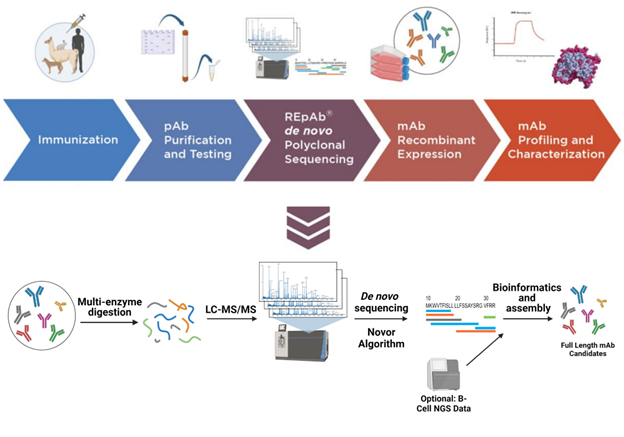
Image Credit: Rapid Novor Inc
How does antibody discovery with polyclonal antibody sequencing address some of the main challenges with antibody discovery campaigns that we have discussed?
Proteomics and mass spectrometry-based antibody discovery effectively address several challenges faced by other discovery technologies.
One of the primary advantages of REpAb polyclonal antibody sequencing is its ability to expand the search space by leveraging the native immune system. Obtaining peripheral blood samples is the only observable interface between germinal centers and bone marrow. Utilizing a proteomics-based discovery method to interrogate serum immunoglobulins provides the ability to analyze antibodies secreted directly from the bone marrow. These clones are difficult to find with conventional methods, such as B cell sequencing, where it is impossible to collect a sample that is representative of the immune diversity due to the nature of B cell differentiation.
If you’re looking for your friend, is it easier to find them walking down a random street? Or by knocking on their door? Instead of searching for needles in haystacks, use a magnet. This is what it's like searching for antibodies directly from the source by using a proteomics-based approach on serum.
Polyclonal sequencing is not impacted by the limitations of retrieved B cells or phage libraries and thereby analyzes the entire antibody repertoire at any given time. Consequently, this approach allows for the analysis of diverse antibody repertoires and can identify candidates with desired functionality and specificity that may otherwise be missed.
Image Credit: ShutterStock/ustas7777777
REpAb polyclonal antibody sequencing allows for the identification of antibody candidates with biological relevance. Immunoglobulins present in serum have already been selected by the host's immune system through extensive affinity maturation and somatic hypermutation, yielding functional antibodies. As these immunoglobulins have been generated by the native host in high abundance, there is generally less concern with production yield, post-translational modifications, stability, immunogenicity, and aggregation propensity. However, thorough biophysical and performance characterization is still essential. Our proprietary wet-lab approaches employ methods for positive and negative selection during affinity purification protocols to zero in on functional binders against specific epitopes of interest. We employ several strategies to yield biologically relevant binders.
REpAb polyclonal antibody sequencing utilizes a de novo approach, deriving the amino acid sequence directly from the MS/MS spectra without reference to known protein or DNA sequences. This species agnostic antibody discovery approach proves to be beneficial for several reasons, including the unique advantage in using different hosts to raise antibodies against conserved proteins. Antibodies have high sequence diversity due to extensive somatic hypermutations during in vivo affinity maturation processes. As such, sequencing antibodies de novo is the most effective method, as often reference sequences are not available. Additionally, it offers a valuable discovery technology when other methods, such as hybridoma generation, are not feasible for certain species.
Where can readers find more information about your company’s technologies?
About Dr. Xiaobing Han
Dr. Xiaobing Han brings 20 years of experience in immunological research, with a specific focus on antibody therapeutics development, for 14 years. At Rapid Novor, Xiaobing leads the antibody discovery team, conducting investigations into antibody responses against a diverse range of targets across various species.