What challenges do researchers usually encounter with conventional proteomic methods?
The proteomics field is highly complex, spanning initial biological samples to comprehensive analysis. An overarching theme repeatedly emphasized by researchers revolves around the absence of a universal solution for this entire process. Rather, each phase demands meticulous harmonization.
Researchers often find themselves at a crossroads. They must choose between targeted approaches, such as arrays, immunoassays, or gels, where the search is predefined, or opt for an unbiased technology that enables in-depth sample interrogation.
Attaining peptide-level resolution predominantly involves mass spectrometry — a costly avenue, with machines exceeding a million-dollar price tag, accompanied by demanding staffing and infrastructure prerequisites that not all institutions can accommodate.
How does next-generation protein sequencing provide a unique solution for these challenges?
In considering this question, it is essential to recognize two distinct customer groups, each uniquely impacted by the advantages of next-generation protein sequencing.
For someone dedicated to overseeing a core lab within a well-funded academic research institute, catering to advanced technologies, including mass spectrometry and others, I think this technology allows experts to delve deeper into their sample analyses.
While initial screening reveals the composition of the samples, next-generation protein sequencing technology facilitates a comprehensive interrogation of amino acid variations, modifications, and intricate changes that solely sequencing can unveil, scrutinizing peptides at the amino acid level.
Conversely, there are also some researchers who may lack the extensive infrastructure or financial means to procure costly mass spectrometry equipment. Presently, they outsource their sample analyses to core labs.
This is where the pivotal aspect of accessibility comes into play. Our proprietary machine, available at an affordable cost of $70,000, grants them the autonomy to conduct testing within their laboratories.
The integration of automated analysis negates the necessity for specialized personnel skilled in tasks like bioinformatics. For this group, the focus shifts towards internalizing and maintaining control over their research process, reducing reliance on external core labs.
Can you explain how the Platinum® Next-Generation Protein Sequencer makes protein sequencing more accessible to researchers from varying scientific backgrounds?
Platinum® Next-Generation Protein Sequencer achieves this through two key avenues. Firstly, it addresses the financial aspect. Irrespective of whether an institution is large or small, acquiring such equipment demands a budget allocation.
Priced at $70,000, this instrument removes the necessity of securing substantial grants solely to access this technology. Researchers can more easily identify resources to make this technology investment, facilitating its integration into their research endeavors.
Secondly, the ease of data analysis is exceptional. While certain analyses, such as protein identification, can be performed on both our machine and a mass spectrometer, our platform distinguishes itself through automated post-analysis procedures.
In contrast, with a mass spectrometer, you either have to have the staff to help conduct this analysis, or you would have to spend the time to do that analysis yourself. Platinum® Next-Generation Protein Sequencer not only ensures simplicity but also alleviates the onus on specialized staff, making routine tasks like protein identification more efficient.
How does the Platinum sequencer provide high-resolution protein sequence data without requiring extensive expertise or complex infrastructure, and why is this important?
The fundamental principle underlying this technological approach is the utilization of standard sample preparation methods commonly employed in proteomics laboratories.
These labs typically utilize techniques such as immuoprecipitation or sample depletion methods. The innovative aspect of next-generation protein sequencing, which drives enhanced insights, revolves around the breakdown of proteins into numerous individual peptides.
This technology employs a combination of specialized sequencing reagents, a semiconductor chip, and a dedicated machine. This combination facilitates a careful process wherein the peptide is sequentially cleaved, one amino acid at a time, and accurately measured.
Consequently, this procedure's intricacy and labor-intensive nature, which would conventionally demand significant resources within a laboratory setting, are efficiently condensed within our proprietary machine and technology. The culmination of this process yields invaluable data.
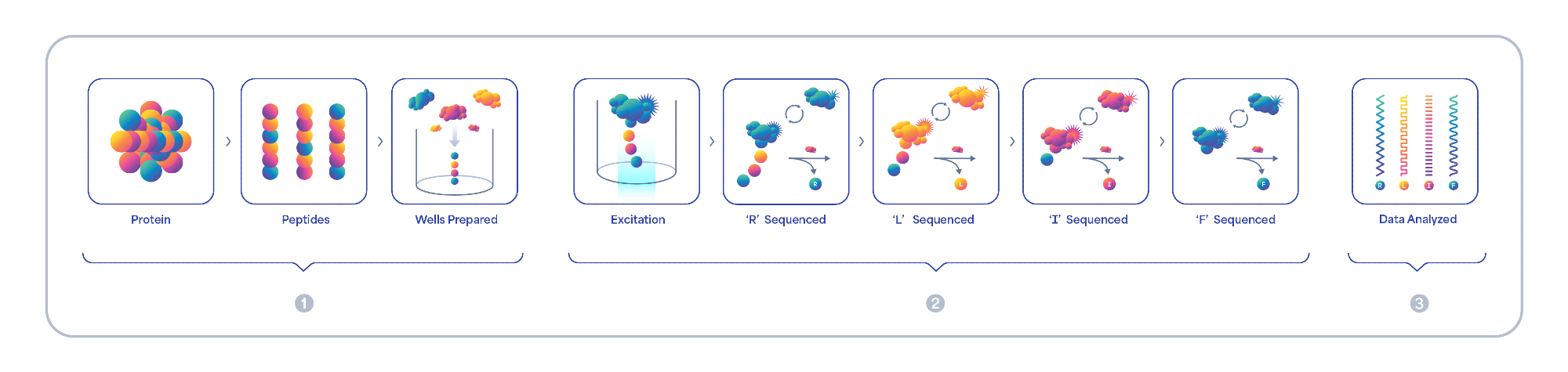
Figure 1. Platinum’s easy-to-use, end-to-end solution includes everything you need to 1) prepare, 2) sequence and 3) analyze proteins with seamless integration into existing workflows. The Platinum workflow begins with protein digestion and immobilization of peptides on a semiconductor chip. Fluorescently labeled N-terminal amino acid (NAA) recognizers bind each NAA and the binding intensity and kinetics are captured as a unique kinetic signature for each NAA. Aminopeptidases cleave NAAs exposing the next NAA for sequencing. On-off binding reveals the amino acid sequence in each diverse peptide. Data is securely transferred and analyzed using an intuitive Cloud based software which aligns and translates kinetic signatures to protein identification.

 Discover the Platinum® Next-Generation Protein Sequencer
Discover the Platinum® Next-Generation Protein Sequencer
How does next-generation protein sequencing complement existing workflows, especially for researchers primarily focused on genomics or transcriptomics?
The significant advantage of next-generation protein sequencing lies in its capacity to delve deeper into samples, down to the amino acid level — an aspect that sets it apart from many other technologies. Whether you operate within a proteomics core or a genomics core, this capability holds substantial value.
Transitioning from genomics to proteomics introduces considerations of accessibility and cost, such as the expenses associated with automated analysis machines and specialized capabilities not typically found in genomics cores.
Fundamentally, genomics laboratories are embracing proteomics technologies to complete the entire narrative. Their journey begins with DNA-level work, extends to RNA and transcriptomics, and ultimately seeks to unveil the intricacies of protein presence and expression. The objective is to establish a cohesive connection between RNA sequencing outcomes and observations at the proteomic level.
Researchers aim to ascertain whether proteins align with anticipated results in terms of their presence and functionality across all subjects or patients under scrutiny. This pursuit involves investigating potential deviations — be it the absence of expected proteins or the non-uniform performance of identified proteins among subjects.
Researchers also strive to uncover protein modifications and diverse proteoforms, further enriching the multiomic exploration. The progression spans from foundational DNA analysis to transcriptomics and culminates in a comprehensive understanding of functional occurrences at the protein stratum.
What valuable insights can researchers gain from the Platinum® Next-Generation Protein Sequencer?
There are distinct customer segments that derive unique benefits from Platinum® Next-Generation Protein Sequencer. One group comprises individuals who seek to conduct their research autonomously, thereby exerting control over the research timeline and maintaining ownership from sample collection to eventual publication. For them, this technology fulfills the fundamental aspiration of comprehensive research ownership across all stages.
Conversely, those already immersed in proteomics harness the capabilities of the sequencer to delve into the intricacies of information. In cases where samples harbor hypothesized amino acid alterations or discrepancies between responder and non-responder profiles, this tool facilitates the investigation of potential protein modifications, variants or other changes.
Researchers at this point possess hypotheses that they may have pursued using existing methods, yet they aspire to delve even further, pinpointing underlying causes and substantiating their conjectures. The sequencer empowers them to embark on a journey of discovery, enabling the exploration of factors they consider causative or significant yet are unable to accurately detect using current methodologies.
How does the Platinum® Next-Generation Protein Sequencer enhance the capabilities of researchers using immunoassays for protein analysis?
When utilizing Platinum® Next-Generation Protein Sequencer in conjunction with immunoassays for protein analysis, a significant shift in approach becomes apparent. In the realm of immunoassays, the conventional process involves deliberate selection and targeting of specific proteins, a biased methodology necessitating a clear predefined objective.
In contrast, the distinct advantage offered by Platinum® Next-Generation Protein Sequencer lies in its capacity for unbiased analysis. Within the domain of proteomics, this technology permits researchers to explore samples without the constraint of predefining their target. This attribute fundamentally alters the dynamics, rendering the traditional necessity of repeatedly procuring or designing specialized reagents obsolete.
Each iteration of a study in the traditional immunoassay paradigm necessitates the acquisition or creation of specific reagents tailored to the protein of interest.
In contrast, Platinum® Next-Generation Protein Sequencer streamlines this process by enabling uniformity in workflow and reagent usage across diverse investigations. This consistency is maintained regardless of the specific protein being studied, offering unparalleled efficiency and resource utilization.
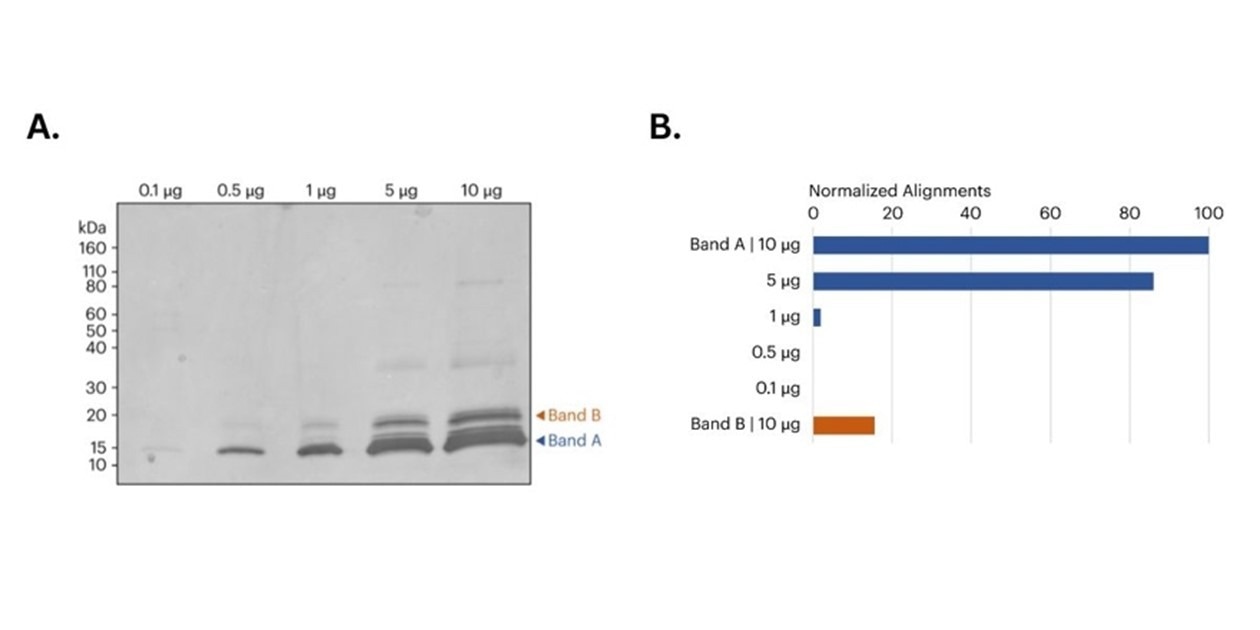
Figure 2. A) image of SDS-PAGE gel of CDNF samples prior to band excision. Orange and blue arrows indicate the approximate location of the bands removed for in-gel digestion and subsequent protein sequencing on Platinum. B) Bar graph shows the relative number of peptide alignments for each in-gel digested sample. For Band A, both the 5 μg and 10 μg samples produced more aligned CDNF peptides compared to the 0.1 μg, 0.5 μg, and 1 μg samples. Additionally, the 10 μg Band B sample also produced a significant number of CDNF aligned peptides. All alignments normalized to the total alignments from the 10 μg library.
The inherent advantage of employing an unbiased technology is the capacity to not only identify the sought-after protein but also uncover additional proteins of interest that might have gone otherwise unnoticed.
This capacity is particularly crucial when piecing together a comprehensive understanding of protein interactions and dynamics within a complex biological system.
Can you share specific examples where next-generation protein sequencing has significantly advanced proteomics research and improved our understanding of complex biological processes?
A compelling example, detailed in an application note available on our website, stems from a collaborative effort with an academic institution. In this case, the focus was on individuals who had contracted COVID-19. The aim was to discern variations in the immune responses among those infected by different strains like Omicron, Delta, and the original Alpha strain, thereby unraveling crucial distinctions.
Through this partnership, we managed to unveil subtle alterations, such as single amino acid changes and the presence/absence of specific peptides. The ability to sequence at the amino acid level played a pivotal role. By leveraging this approach, it became feasible to accurately pinpoint whether an individual was impacted by Omicron or Delta, effectively enabling a form of surveillance.
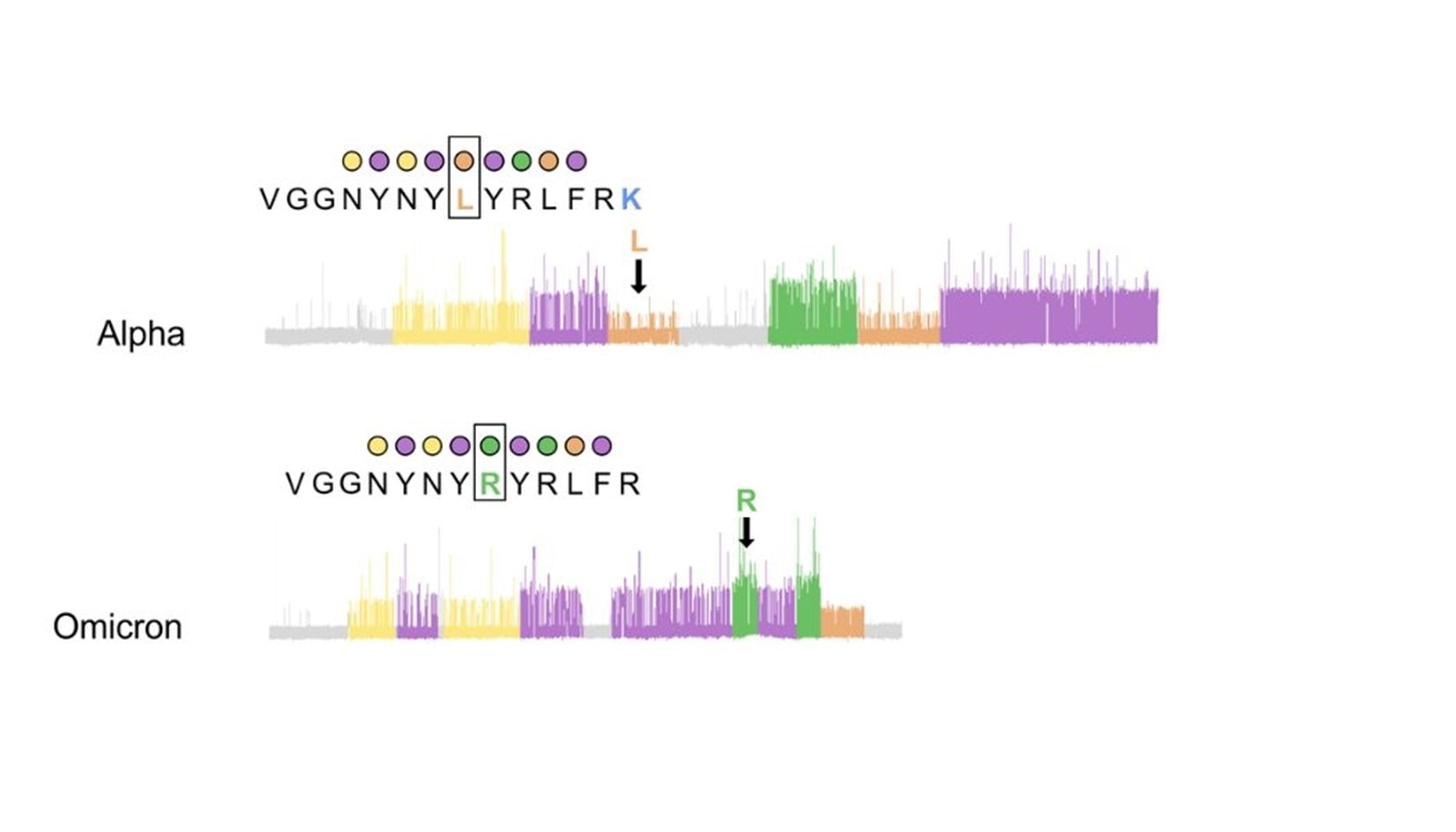
Figure 3. Example protein sequencing traces of SARS-CoV-2 variants for the peptide where the L452R mutation occurs shows that recognizer binding produces clear kinetic differences influenced by the L452R mutation that can be detected with Quantum-Si’s Platinum instrument, effectively differentiating the Alpha variant from Omicron.
The significance lies in the scenario where individuals had encountered COVID-19 but the viral RNA had already dissipated. This method offered a unique avenue to investigate the aftermath of infections and analyze the immune responses that persisted.
Essentially, it allowed for the monitoring of individuals' immune reactions post-infection. This avenue of research holds particular intrigue since it extends the study timeline well beyond the phase when viral RNA can be sequenced, thereby facilitating the identification of viral strains even after their genetic material has waned.
What advantages does the Platinum sequencer offer over traditional mass spectrometry for identifying proteins and proteoforms?
Regarding protein identification, mass spectrometry excels in scenarios where you need to process a large number of samples in one go — say, working with 100 or 50 samples simultaneously.
However, it is important to note that not every research endeavor requires such extensive scalability. There are instances when your sample pool is smaller or you are seeking a swift and streamlined identification process without navigating the complexities associated with a high-volume workflow.
Conversely, when the objective is to delve deeper into analysis, scrutinizing protein variants becomes pivotal — encompassing protein modifications and single amino acid substitutions. While certain mass spectrometry methods can address some of these intricacies, they often entail arduous preparatory stages and other procedural intricacies.
This is where Platinum sequencer distinguishes itself by enabling the execution of these analyses through a more standard workflow on our platform. The distinct advantage emerges as you aim for profound insights, delving into intricate protein details.
However, it is worth highlighting that there is also value in conducting fundamental protein analyses, especially before your research undertakings advance to the scale of hundreds of samples. In these contexts, Platinum offers a swifter and more accessible approach to protein identification.
How do you see next-generation protein sequencing shaping the future of proteomics research and contributing to scientific discoveries in this field?
The significant impact of next-generation protein sequencing lies in its potential to drive transformative discoveries. An array of instances involving targeted therapeutics underscores the relevance of pinpointing specific proteins.
Upon analyzing clinical data, patterns emerge where individuals exhibit distinct responses, including responders and non-responders, as well as recurrence and non-recurrence cases. Delving into this divergence and elucidating its underpinnings constitutes a focal point of exploration for numerous researchers and publications.
The foundation of these disparities often relates to post-translational modifications (PTMs) and a broader spectrum of protein variations, which, while fundamentally sharing protein-level similarity, diverge at the amino acid or PTM level.
This divergence offers a valuable avenue for stratification, enabling refined identification and the advancement of targeted treatments. Moreover, this technology bears the potential for patient stratification, diagnostics, and therapeutic assessments. It could serve as a disease biomarker or aid in monitoring responses, even detecting minimal residual disease.
Notably, both diagnostic and therapeutic realms stand to gain from the newfound capacity to unveil these intricacies. The prevailing scenario of uniform treatment yielding diverse outcomes prompts an unbiased, comprehensive exploration to uncover the underlying factors.
This understanding holds the promise of novel therapeutic approaches, individualized treatments, and early intervention strategies. Ultimately, the insights achievable through such comprehensive analysis pave the way for long-term advancements in proteomics research, ushering in a new era of potential breakthroughs.
In what unique ways does the Platinum sequencer allow researchers to study modifications, proteoforms, and rare events that antibodies may not detect?
Platinum sequencer offers several distinct advantages in this regard. Our technology presents a more versatile approach than relying solely on antibodies, which necessitates prior knowledge of the specific change being targeted.
Researchers often face limitations when using antibodies, particularly when dealing with subtle modifications within a protein. The intricacy of these modifications could render it challenging to develop highly specific antibodies tailored to these minute alterations.
However, with Platinum next-generation sequencing, this challenge is mitigated. By enzymatically breaking down the protein into peptides and subsequently sequencing them at the amino acid level, our technology empowers researchers to explore without the need for specialized reagents.
This means that researchers no longer have to develop new reagents or precisely know in advance what to search for. The sequencer facilitates a broader lens approach, allowing researchers to uncover a spectrum of alterations — be it probing for specific protein modifications or identifying substitutions — without the constraints of traditional antibody-based methods.

Image Credit: Quantum-Si
How has the Platinum® Next-Generation Protein Sequencer encouraged researchers to adopt a more comprehensive multiomics approach?
In the early stages of our commercial launch, we observed a notable trend among researchers. Specifically, at the proteomics core laboratory level, there is a growing enthusiasm for conducting in-depth analyses as part of their research endeavors.
This enthusiasm arises from the desire to meticulously decipher distinctions between diverse study populations. As discussed earlier, our presence at trade shows and similar events, often with a genomic focus, has allowed us to witness an increasing number of genomics labs aiming to enrich their capabilities as well.
These labs have already delved into DNA and RNA sequencing, and some are even venturing into spatial biology domains.
The introduction of proteomics, the capacity to sequence proteins, serves as a valuable supplementary tool for their holistic exploration. This approach empowers researchers to investigate their areas of interest comprehensively, spanning from the DNA level to the functional dimension of proteins.
About Jeff Hawkins
 Jeff Hawkins has over 20 years of experience at the world’s leading life science and diagnostics companies.
Jeff Hawkins has over 20 years of experience at the world’s leading life science and diagnostics companies.
Prior to Quantum-Si, Jeff was President and Chief Executive Officer of Truvian Sciences, Inc. where he led the evolution of the company’s benchtop blood testing system from a product concept. Jeff Hawkins holds a B.A. in Chemistry with honors from Concordia University as well as an MBA from Keller Graduate School of Management.
About Quantum-Si
 Quantum-Si was originally founded in 2013 by Jonathan Rothberg, who envisioned a new frontier where biologists can accelerate basic scientific insights and biomedical advances through the power of next-generation protein sequencing.
Quantum-Si was originally founded in 2013 by Jonathan Rothberg, who envisioned a new frontier where biologists can accelerate basic scientific insights and biomedical advances through the power of next-generation protein sequencing.
Quantum-Si aims to revolutionize proteomics, by bringing single-molecule protein analysis to every lab, everywhere, to generate a better understanding of proteins' role in cell biology and disease. The company’s instrument, called Platinum®, is powered by a first-of-its-kind semiconductor chip, and enables researchers through next-generation protein sequencing to understand what is truly happening at the protein, peptide or amino acid level to fuel new discoveries.