 Study: Gut microbiome is not associated with mild cognitive impairment in Parkinson’s disease. Image Credit: CI Photos / Shutterstock
Study: Gut microbiome is not associated with mild cognitive impairment in Parkinson’s disease. Image Credit: CI Photos / Shutterstock
About the study
The present study investigated whether gut microbes are related to mild cognitive impairments in PD patients.
The researchers analyzed the Luxembourg PD Study data to compare the gut microbial characteristics of 58 Parkinson's disease patients with mild cognitive impairments (PD-MCI group), 60 cognitively unimpaired Parkinson's disease patients (PD-NC group), and 90 control individuals with normal cognitive development. The study participants matched the United Kingdom PD Society Brain Bank clinical diagnostic criteria for PD.
The team defined MCI using the Movement Disorder Society (MDS) guidelines. They used the Montreal Cognitive Assessment (MoCA) and Unified PD Rating Scale (MDS-UPDRS) for cognitive evaluation in PD. They used the Rome III criteria to ascertain constipation and previously published conversion factors to calculate the levodopa equivalent daily dosage (LEDD). They used the DADA2 tool for quality filtering, chimera removal, amplicon sequence variant (ASV) generation, and differential abundance comparisons.
The team excluded individuals with atypical or not yet specified parkinsonism, controls genetically related to PD participants, and individuals with missing data on covariates such as body mass index (BMI) or educational attainment. They also excluded individuals reporting corticosteroid or immunosuppressant use in the previous six months and those with less than 10,000 sequence reads remaining after removing rare ASVs and mitochondria and chloroplast ASVs. As a result, the sample dataset included 208 individuals.
Results
The control group had a lower age and a reduced constipation frequency, while the PD groups exhibited comparable characteristics. There were no changes in gut microbial community evenness (or alpha diversity) or richness across the research groups after confounder exclusion. The linear regressions for inverse-type Simpson index values, comprising the PD-MCI, PD-NC, and control groups with possible confounders, revealed that the PD-MCI and PD-NC groups showed fewer diversities than controls. Within-PD modeling with confounding factors revealed no difference in the PD-MCI and PD-NC groups.
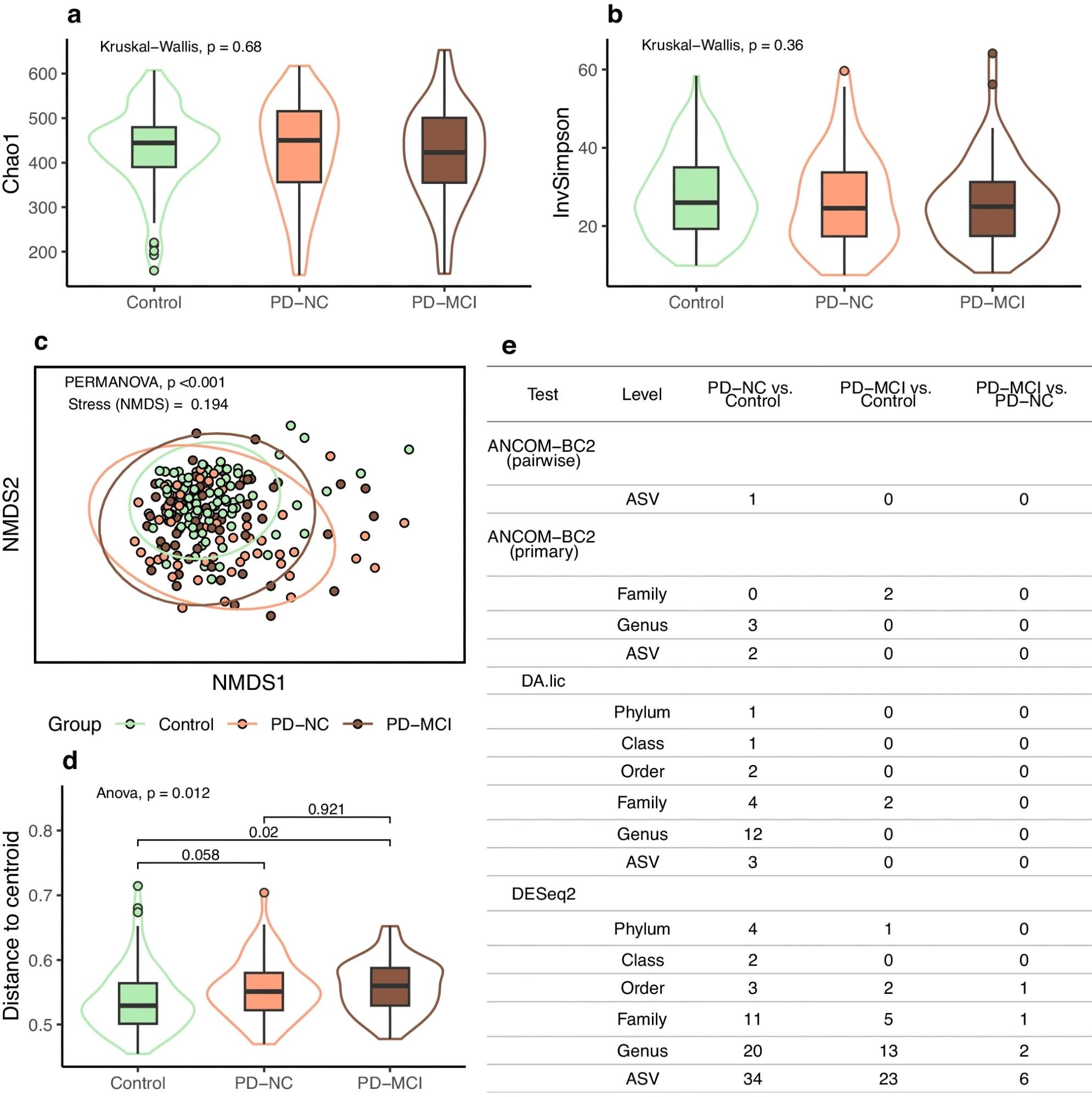 a Boxplot for richness (Chao1). b Boxplot for richness and evenness (inverse Simpson). c Community composition visualized as NMDS ordination of Bray-Curtis dissimilarity; ellipses indicate 95% confidence intervals. d Boxplot for groupwise distances to centroid from the ordination, with significances for pairwise comparisons from Tukey HSD test. e Numbers of differentially abundant taxa (multiple comparison corrected p < 0.05). In boxplots, box hinges represent the 1st and 3rd quartiles, whiskers range from hinge to the highest and lowest values that are within 1.5*IQR of the hinge, and outlines represent data distributions.
a Boxplot for richness (Chao1). b Boxplot for richness and evenness (inverse Simpson). c Community composition visualized as NMDS ordination of Bray-Curtis dissimilarity; ellipses indicate 95% confidence intervals. d Boxplot for groupwise distances to centroid from the ordination, with significances for pairwise comparisons from Tukey HSD test. e Numbers of differentially abundant taxa (multiple comparison corrected p < 0.05). In boxplots, box hinges represent the 1st and 3rd quartiles, whiskers range from hinge to the highest and lowest values that are within 1.5*IQR of the hinge, and outlines represent data distributions.
The comparison of community compositions (beta diversity) revealed variations between the research groups, regardless of confounders. Pairwise testing between control individuals and PD patients revealed significant group effects, while the within-PD analysis found no changes in MCI status. Sample dispersion testing showed significant differences between PD patients with mild cognitive impairments and controls, almost statistically significant differences between the PD-NC group and controls, and no significant differences between the PD-NC and PD-MCI groups.
Taxa that were significant across several tests included lower abundances of Butyricicoccaceae, Lachnospiraceae, and Clostridiaceae in PD, as well as elevations in Enterobacteriaceae, Ruminococcaceae DTU089, and Hungatella. DESeq2 analysis revealed increases in Shigella, Methanobrevibacter, and Escherichia coli. However, PD-MCI and PD-NC group comparisons yielded non-significant results in two of three assessments. DESeq2 identified ten significant microbial taxa, significantly lower than in other group comparisons. Streptococcus (raised among PD-MCI patients) and Akkermansia muciniphila ASV (decreased in the PD-MCI group) were two of the most significant taxa.
Conclusions
The study found no particular microbiome signatures associated with mild cognitive impairment in Parkinson's disease. Many taxa were differently abundant across PD groups and controls, with PD having higher abundances of Methanobrevibacter, Enterobacteriaceae, and Hungatella and lower abundances of Butyricicoccaceae and Lachnospiraceae. Beta diversity findings were also consistent with earlier research.
There were no statistically significant beta diversity differences among PD-MCI and PD-NC individuals. The absence of overlap supports a consistent microbiological profile for mild cognitive impairment in Parkinson's disease. The most intriguing taxon discovered was an Akkermansia muciniphila ASV, almost missing in the PD-MCI group. Further research is required to determine the role of Akkermansia muciniphila in PD subtypes.
Journal reference:
- Aho, V.T.E., Klee, M., Landoulsi, Z., et al. The gut microbiome is not associated with mild cognitive impairment in Parkinson's disease. npj Parkinsons Dis. 10, 78 (2024), DOI: 10.1038/s41531-024-00687-1, https://www.nature.com/articles/s41531-024-00687-1