The 3View® system is an advanced three-dimensional electron microscopy (3DEM) system that has been designed to conduct serial block-face scanning electron microscopy (SBFSEM).
The new generation scanning electron microscope (SEM) has extended the performance limits of 3DEM in research as well as in clinical fields.
In the Journal of Electron Microscopy, a research group from the School of Life Sciences and Biotechnology at Korea University has validated the effects of laser treatment for melisma using Gatan 3View data.
In the paper, the researchers described their search for melanosomes and the creation of 3D models of melanocytes using the Gatan 3View data. The study results have been reported on the cover of the journal.
Study results
From the breakthrough research carried out by Winfred Denk in MPI Heidelberg, 3D ultrastructure can be automatically acquired using the 3View system. Here, a freshly cut, resin-embedded block-face is sequentially imaged.
An ultramicrotome is placed within the SEM and the material is removed using a diamond knife to expose a new surface for subsequent imaging. An embedded sample can be turned into thousands of images overnight, thus eliminating the need for cutting and collecting ultrathin sections.
The images collected are arranged in a line at the point of acquisition as opposed to serial section transmission electron microscopy (TEM). The 3D image data can be viewed and browsed by the users in real time during the acquisition.
Melasma, a common dermalogical skin disease, involves the appearance of a dark skin discoloration on areas of the face upon sun exposure. The structural changes of melanocytes were studied by J.Y. Mun et al after subjecting them to Q-switched Nd:YAG laser exposure using SBFSEM (Figure 1). Melanocytes are present in the bottom epidermis layer and their dendrites can be found across all epidermis layers (Figure 2).
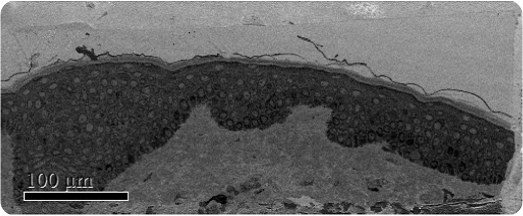
Figure 1. Low magnification SBFSEM image displaying the entire block-face sample with a field of view of 200 x 500μm.
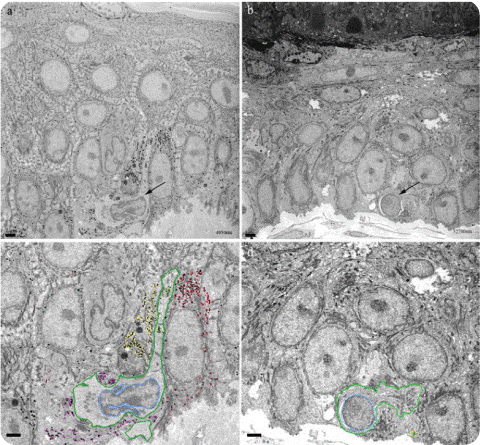
Figure 2. SBFSEM images of the epidermis layer. (A) and (C) images of pre-laser treatment. (B) and (D) images of post-laser treatment. The images (A) and (B) show one section among over 500 serial sections. Arrow in (A) and (B) represent melanocytes. In (C) and (D), dark green represents melanocytes and light blue show nucleus. Yellow, purple, red and cyan dots show melanosomes. The images (C) and (D) show a decrease of the volume of melanocyte and the number of melanosome after laser treatment. Size bar is 2μm.
The dendrites of melanocytes cannot be resolved by 3DEM as they are very small and often too deeply located. However, the 3View system allows for slicing down to an area of interest, unhindered by structures that are too deep to be resolved by confocal or two-photon microscopy.
In this study, 500 serial images were acquired by the 3View system, using 50nm isotropic voxel size and a 28 x 28μm field of view (Figure 3). The data acquisition with isotropic voxels enables viewing the data from any orientation with no distortion (Figure 4).
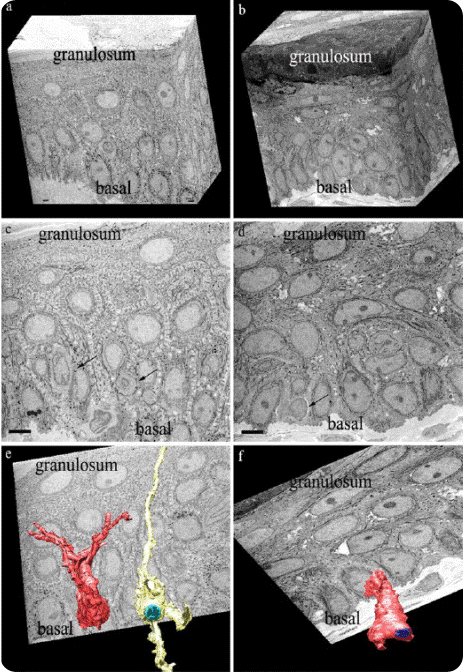
Figure 3. Shows the epidermis layer of patients with melisma, and the 3D melanocyte structures. (A), (C) and (E) images of pre-laser treatment and (B), (D) and (F) images of post-laser treatment. 3D volume of over 500 serial images using the 3View system was visualized by IMOD (E) and (F). (C) and (D) one of the serial images for 3D reconstruction of (E) and (F). The size bar of (C) and (D) is 5μm. The images (E) and (F) show the surface model of melanocytes showing cell body and their dendrites. Each nucleus was assigned a separate color (E, cyan blue; F, blue). Many dendrites are present in the melanocytes of melasma patients, which stretch from granulosum to basal (E). After laser toning treatment, the 3D structure of melanocytes contained fewer dendrites (F).
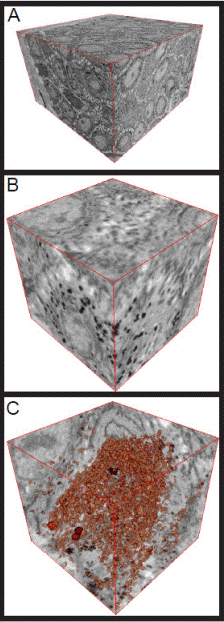
Figure 4. Raw data displayed with the 3D visualization tool in the DigitalMicrograph software. (A) is a 3D visualization of the complete dataset displaying the complete volume. (B) is a 3D extract of displaying a specific region of interest. (C) is the same region of interest thresholding out the melanosomes in red and displaying ortho-slices in all three planes.
Conclusion
In comparison with serial section TEM throughput, high-resolution volumes that are previously unfeasible with traditional techniques are collected by the 3View system. A 100 – 200μm3 dataset can be collected within a period of 24 hours by the 3View system, using 50nm isotropic voxels.
The 3View system can resolve subcellular structures like cristae within the desmosomes and mitochondria. Large fields of view can also be covered by the 3View system by applying the stage montage feature available in the DigitalMicrograph® software.
About Gatan Inc.

Gatan, Inc. is the world's leading manufacturer of instrumentation and software used to enhance and extend the operation and performance of electron microscopes.
Gatan's products, which are fully compatible with all brands of electron microscopes, cover the entire range of the analytical process from specimen preparation and manipulation to imaging and analysis.
Our customer base spans the complete spectrum of end users of analytical instrumentation typically found in industrial, governmental and academic laboratories.
The applications addressed by these scientists and researchers include metallurgy, semiconductors, electronics, biological science, new materials research and biotechnology.
The Gatan brand name is recognized and respected throughout the worldwide scientific community and has been synonymous with high-quality products and the industry's leading technology.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.