Reliable assays are a vital factor in ensuring timely sample processing during SARS-CoV-2 testing. It is important to mitigate potential delays along the workflow in order to improve quality and ensure reliable viral detection via reverse transcription quantitative PCR (RT-qPCR).

Thermo Scientific NanoDrop One/OneC Microvolume UV-Vis Spectrophotometer. Image Credit: Thermo Fisher Scientific – Materials & Structural Analysis
The Acclaro™ Sample Intelligence technology built into the Thermo Scientific™ NanoDrop™ One Microvolume UV-Vis Spectrophotometer provides users with a robust toolset that ensures accurate nucleic acid concentrations, better identifies contaminants and helps avert experiment failure.
Testing considerations for SARS-CoV-2 samples
The World Health Organization (WHO) published the genetic sequence for SARS-CoV-2 - the virus that causes the COVID-19 disease - in early 2020. By 11 March 2020, the disease had spread to the point that it was classified as a pandemic.
By 3 August 2020, more than 18 million individuals around the world had tested positive for the virus. Patient testing has been the primary focus for government organizations, private laboratories and hospitals.
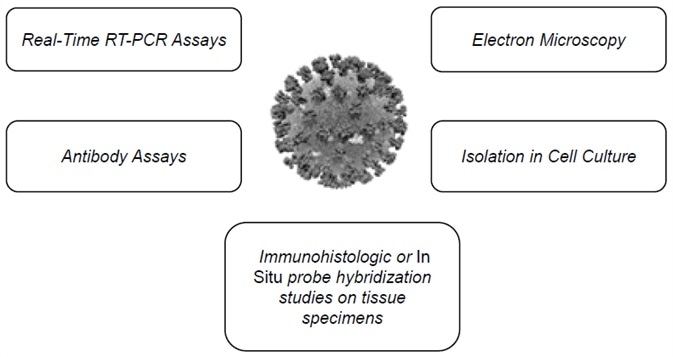
Figure 1. Multiple SARS-CoV-2 detection workflows. Image Credit: Thermo Fisher Scientific – Materials & Structural Analysis
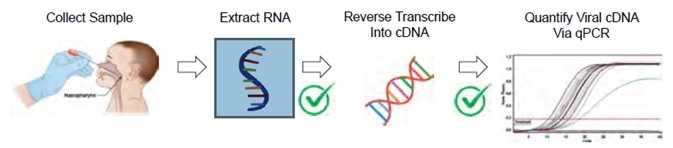
Figure 2. One SARS-CoV-2 workflow is a Real-Time RT-qPCR Assay. This figure is adapted from the CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel2. Note that the CDC guidelines do not mention nucleic acid quantification after RNA extraction or before qPCR amplification. The MIQE guidelines for minimum information for publication of quantitative real-time PCR experiments3 do define nucleic acid quantity and A260/A280 purity as useful information for qPCR publication. Image Credit: Thermo Fisher Scientific – Materials & Structural Analysis
The Centers for Disease Control and Prevention outlines several assays suitable for the detection of the virus,1 including RT-qPCR, electron microscopy, antibody assays and more (Figure 1 and Figure 2).
In Q2 2020, emergency use authorization was awarded to the Applied Biosystems™ TaqPath™ COVID-19 Combo Kit and a number of qPCR instruments by the U.S. Food and Drug Administration.4
Importance of nucleic acid quantification
The development of reliable RT-qPCR assays requires a combination of thorough experimental design, extensive QC and transparent data analysis. Leaders in the qPCR field shared the MIQE guidelines in 2009, aiming to improve experiment quality and reliability.
A typical RT-qPCR assay features two distinct enzymatic steps:
First, RNA is converted to cDNA via the use of reverse transcriptase. This normalizes the quantity of RNA going into the reaction, minimizing variability in cDNA production. This is a critical consideration due to the potential for RT reaction efficiencies to change with varying RNA concentrations.
The second step sees gene-specific primers utilized in combination with cDNA templates to ascertain the relative expression versus various reference genes.
Reference genes are a key element of this reaction due to their role in normalizing small differences in cDNA input. Large differences in the cDNA amount, normalization based on reference genes can become challenging. MIQE guidelines require input RNA quantity and purity to be reported.
Using the NanoDrop One Spectrophotometers for qPCR workflows
The NanoDrop One Microvolume UV-Vis Spectrophotometer is widely regarded by global scientists in the quantification of small volumes of nucleic acid samples. These instruments feature the Acclaro Sample Intelligence technology, which can identify and quantify the concentration of many common contaminants in samples.
Molecules frequently encountered in nucleic acid extraction kits can cause an overestimation of analyte concentrations or trigger the denaturing of qPCR polymerases, ultimately causing a qPCR experiment to fail.
The Acclaro software is able to provide corrected nucleic acid concentrations and can identify the majority of common contaminants, helping minimize the time required to troubleshoot and address failed experiments – an essential consideration when aiming to mitigate these issues under pandemic timelines.
Results
A number of CDC-approved nucleic acid extraction kits utilize guanidine thiocyanate (GuSCN) and guanidine HCl in their chemistries.
The presence of GuSCN contamination in a sample will not significantly alter reported absorbance values at 260 nm or calculated nucleic acid concentrations, but this can denature qPCR polymerases.
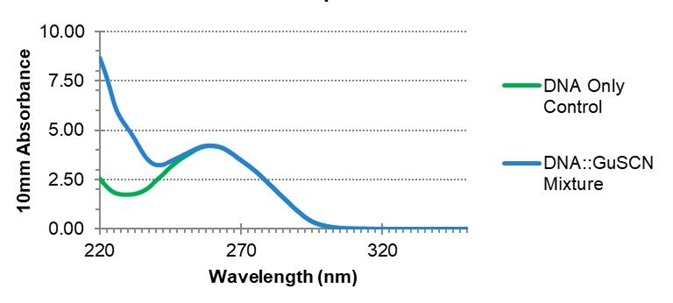
Figure 3. The effect of guanidine thiocyanate on nucleic acid spectra. The absorbance at 260 nm is unaffected by the presence of GuSCN, but the A260/A230 ratio may be impacted. Image Credit: Thermo Fisher Scientific – Materials & Structural Analysis
To illustrate this issue, RNA was extracted from a mouse liver and spiked with a GuSCN contaminant (Figure 3). Absorbance at 260 nm was used to ascertain the analyte concentration and develop a qPCR based on the uncorrected value.
Guanidine salt contamination has the potential to inhibit qPCR by denaturing polymerases, meaning that its presence could falsely increase the cycle threshold (Ct) count. Ct was measured, confirming that it increased in line with contaminant concentration.
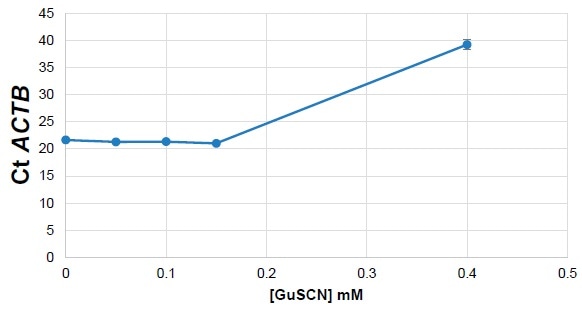
Figure 4. Mouse Liver RNA spiked with GuSCN. Image Credit: Thermo Fisher Scientific – Materials & Structural Analysis
The reaction did not go as planned (Figure 4), and this was attributed to the presence of guanidine salts inhibiting the polymerases and blocking the reaction.

Figure 5. Acclaro technology contamination alerts. Acclaro technology A) flags an alert for a contaminant present in a dsDNA sample with an icon ( ) and B) offers additional details about the protein contaminant in the sample that contributes to A260 that inflates dsDNA concentration. Acclaro provides uncorrected values and corrected values, where the protein contribution is subtracted from the original value to provide the corrected concentration. Image Credit: Thermo Fisher Scientific – Materials & Structural Analysis
) and B) offers additional details about the protein contaminant in the sample that contributes to A260 that inflates dsDNA concentration. Acclaro provides uncorrected values and corrected values, where the protein contribution is subtracted from the original value to provide the corrected concentration. Image Credit: Thermo Fisher Scientific – Materials & Structural Analysis
The Acclaro software built into the NanoDrop One is able to detect GuSCN in RNA samples, offering a range of features that are able to help avert failed qPCR experiments (Figure 5 and Table 1).
Table 1. Contaminant range detectable by Acclaro technology. Source: Thermo Fisher Scientific – Materials & Structural Analysis
| |
dsDNA |
RNA |
| Contaminants detected |
- protein
- phenol
- guanidine HCl
|
- protein
- phenol
- guanidine SCN
|
| Sample concentration range |
0.5A-62.5A at 260 nm |
0.5A-62.5A at 260 nm |
The Acclaro technology flags that a contaminant is present in a dsDNA sample while providing additional details about the protein contaminant contributing to A260 and inflating dsDNA concentration.
Acclaro delivers both uncorrected values and corrected values, whereby protein contribution is subtracted from the original value to determine its corrected concentration.
Conclusions
The Acclaro Sample Intelligence technology helps to improve confidence in sample assessment, identifying frequently encountered contaminants with the potential to impact downstream RT-qPCR analysis.
The software’s algorithms offer corrected DNA or RNA concentration for each sample in under 8 seconds while calculating accurate A260/A280 and A260/A230 purity ratios.
This functionality allows users to limit costly delays stemming from the need to troubleshoot failed experiments, allowing users to understand the original sample better. These features are essential in time-sensitive assays.
References
- https://archive.cdc.gov/#/details?q=Sars%20assays&start=0&rows=10&url=https://www.cdc.gov/sars/guidance/f-lab/assays.html
- https://www.fda.gov/media/134922/download
- https://www.ncbi.nlm.nih.gov/pubmed/19246619
- For Emergency Use Authorization (EUA) only. For prescription use only. For in vitro diagnostic use.
Acknowledgments
Produced from materials originally authored by Patrick Brown and Brian Matlock from Thermo Fisher Scientific.
About Thermo Fisher Scientific – Materials & Structural Analysis
 Thermo Fisher Materials and Structural Analysis products give you outstanding capabilities in materials science research and development. Driving innovation and productivity, their portfolio of scientific instruments enable the design, characterization and lab-to-production scale of materials used throughout industry.
Thermo Fisher Materials and Structural Analysis products give you outstanding capabilities in materials science research and development. Driving innovation and productivity, their portfolio of scientific instruments enable the design, characterization and lab-to-production scale of materials used throughout industry.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.