The holobiont paradigm finally recognizes that macroorganism development and health, like all other natural ecosystems, are influenced by the diverse microbial communities they host. However, mechanistic insights into microbial community assembly and environmental interactions remain rare.
While it is currently predicted that less than 1% of bacterial organisms can be cultured in the lab, meta-omics (particularly metagenomics) has been the most popular microbiology methodology for the past two decades.
They facilitated a considerable number of high-impact discoveries while also raising an even greater number of concerns.
Whether for fundamental research or to provide reasoned solutions, there is a need to move away from empirical methodologies and learn about the mechanisms underlying the observed patterns.1
Consider this example: Various women's (and infant) health issues, such as immunology, fertility, and cancer risk, are linked to diverse vaginal "microbial signatures," but in which way does this association go?
Can we say for certain that vaginal dysbiosis causes endometriosis, or do they both result from something else, such as contraception or nutrition? From there, do we have the ability to modify the microbiota to restore health sustainably?
The development of single microbial cell techniques is projected to increase knowledge as it will:
- Enable single microbial cell omics, allowing for population and subpopulation characterization and community assembly insights.
- Accelerate the discovery of new microbial physiologies by increasing culture library throughput and breaking down cultivability barriers.
This article will discuss both of these aspects.
Looking at solitary microbial organisms to understand microbial communities: A paradox?
It may appear strange that learning about individual cells could help us understand how communities and ecosystems work. However, single microbial cell omics will make a significant contribution to microbial ecology and microbiology in general because (i) it provides access to population and subpopulation diversity and evolutionary patterns, (ii) it addresses some of the main shortcomings of meta-omics, (iii) it provides a complementary perspective on the same object, and (iv) it allows for a finer and more accurate view of physiological networks.2
Single-cell -omics applied to microbes also present significant obstacles, which will be detailed and discussed below.
Individual, population, community, and environmental microbial interactions range from predation to symbiosis, including competition and mutualism (Figure 1). These are complex and interrelated ecological scales.
Meta-omics approaches in microbial ecology range from community to environmental scales. However, they lack the resolution to capture smaller scales along the ecological gradient, such as population and subpopulation scales.
Microbial (sub)populations, such as species and lower taxonomic ranks, are an understudied reservoir of genotypic and phenotypic diversity, and accessing the intra-population level enables fine-scale analysis of evolutionary processes, gene acquisition/loss processes, population emergence, and, ultimately, community assembly.
When used correctly, particularly in terms of sampling approach and sample size, single-cell omics encompass the entire ecological continuum, from the individual to the community, in the same sample, providing the possibility of connecting information from all levels.
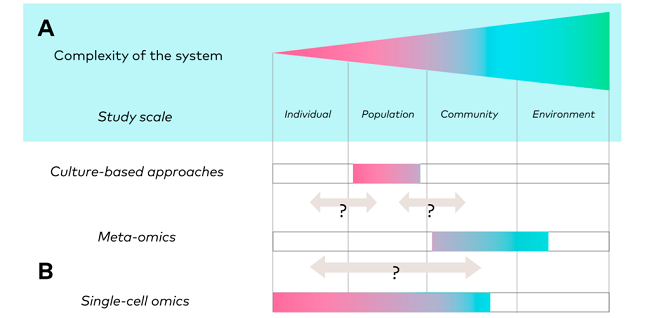
Figure 1. Gradient of ecological scales and associated approaches. Image Credit: Mauger et al. 2021, Trends in Ecology & Evolution 3
Meta-omics generates averaged information from a sample of available molecular information inside an environment.
Drawing population-level insights from meta-omics data remains challenging. During genome reconstruction (metagenomics assembled genomes or MAGs), combining pieces from distinct individual genomes and free DNA might produce chimeras (Figure 2).
MAG accuracy becomes difficult when microbial communities are extremely varied, with closely related taxa having highly similar genomes.
Furthermore, metagenomics cannot distinguish between DNA contained in living cells and free DNA, which can remain in the environment for long periods, or DNA contained in dead cells.4 As a result, meta-omics data cannot directly link a detected gene to its original microbial cell, let alone to its other genes.
Single-cell omics, which is quickly gaining prominence in microbiology, address this issue by radically changing the perspective. It concentrates on the DNA, RNA, or proteins within the same cell, providing new opportunities for researching metabolic pathways or gene networks.
Recent breakthroughs in the field demonstrated that observing microbial communities using metagenomics and single-cell whole genome sequencing (scWGS) provides diverse representations of the diversity within the same samples.5
It implies that the most thorough approach to microbial community composition and function may be to use both meta-omics and single-cell omics on the same sample.
This combines the benefits of both techniques: (i) the ability to assess alpha/beta diversity with high throughput using metagenomics and (ii) the capability for detailed and accurate analysis using scWGS.
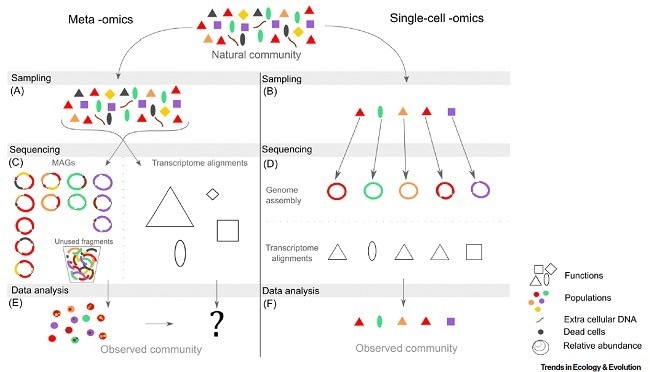
Figure 2. The key steps, features and limitations of meta -omics (left) and single-cell -omics (right). Image Credit: Mauger et al. 2021, Trends in Ecology & Evolution 6
Using single-cell omics to study microorganisms presents significant hurdles.
- Cell isolation requires reliable detection and separation with confirmed monoclonality status. Most isolation methods (including FACS) require fluorescent labeling to effectively detect and successfully isolate (single) tiny cells, which introduces significant biases and is time- and cost-intensive.
- Cells should be put in compartments compatible with molecular biology workflows to prevent loss of molecular material. Additionally, subsequent reactions must be compatible as no purification is possible before cell barcoding.
- Bacterial and archaeal cells contain only a few femtograms of DNA and RNA, approximately 1000-fold less than human cells. DNA and cDNA must be amplified to create sequencing libraries for scWGS and scRNAseq, respectively. Any contamination by foreign DNA (e.g., host DNA), which would be amplified alongside the cell material, could significantly skew the data.7
- High sequencing costs and reliance on library preparation chemicals contribute significantly to overall experimental expenses.
Cellenion believes that cellenONE is the best solution for single-cell omics of microbial cells. It combines high-precision nanolitre-volume liquid dispensing with image-based cell isolation.
- It accurately isolates cells ranging from 0.5 to 80 µm, including microbial, animal, and plant cells (up to 100% for bacteria).
- Cells can be deposited individually in various consumables, including regular multi-well plates and bespoke ones.
- Image-based isolation involves selecting cells of interest based on size, shape, and fluorescence and recording microscope pictures for quality control. It can, therefore, target specific types of cells, such as exclusively prokaryotes, while excluding host cells.
- CellenONE's patented microLIFE software, available since January 2024, achieves maximum accuracy for single-cell isolation even in bright field illumination. Fluorescence labeling is optional.
- The instrument is embedded in a biosafety cabinet (cellenONE BSC), with an inert and sterilizable capillary to prevent contamination and maintain axenic conditions.
- Generates small droplets (100-500 pL) with minimal carry-over liquid, reducing the risk of free DNA contamination.
- It may produce miniature molecular biology reagents as a liquid dispenser, significantly reducing costs.
Single-cell omics may also be a powerful tool for learning more about microbial physiology because it allows access to gene assemblages and route networks at the individual cell level.
It is effective in giving data that allows for the inference of metabolic preferences, which is then utilized to select optimal enrichment conditions, resulting in successful cultivation and pushing the boundaries of microbial dark matter.8
Bring 'microbial dark matter' to light by cultivating the uncultivable
Microbial dark matter is the percentage of microorganisms humans cannot cultivate due to a lack of understanding or control over their growth conditions.
It is widely acknowledged that more than 99% of microorganisms (particularly bacteria and archaea) cannot be cultured in the lab, particularly when they belong to less known (and thus less studied) environments such as soils.9
Molecular investigations demonstrate the ubiquity of taxa revealed through their genetic elements with few to no culture representatives, including several common and prolific ones such as Acidobacteria in soils.10
As a result, microbial physiology research is critically focused on a tiny subset of living microorganisms, and extrapolating culture-based method findings to natural communities is dangerous, if not impossible.
What if we could reduce the amount of microbial dark matter? Let us start anew and reword the first sentence of this section. Microbial dark matter refers to the fraction of bacteria that we are now unable to culture.
Cellenion suggests that (i) boosting culture production throughput is the key to 'microbial brightened matter', and (ii) automated single microbial cell isolation will unlock it.
Traditionally, the most common ways for culture generation are:

Image Credit: Cellenion
- Direct plating (Figure 3A) involves serially diluting a microbe suspension solution and plating it on an agar medium. After the initial incubation, colonies are collected (manually or with a colony picker) and placed in liquid media. Following a second incubation, liquid cultures are often streaked over an agar plate to ensure the purity of the isolated strain. After the third incubation, streaked colonies are chosen and placed in liquid media. After the fourth incubation, cultures can be preserved and/or used for downstream analysis.
- In dilution to extinction (Figure 3B), a microbe suspension solution is serially diluted and injected in liquid cultures (typically in microplates). This method is based on the Poisson law, which states that one well will be inoculated by a single cell. Following an initial incubation, plates containing a high proportion of wells statistically compatible with single-cell inoculation are chosen, and each growing well is streaked for purification at least thrice. After a second incubation, streaked colonies are collected and placed in liquid media. After a fourth incubation, cultures can be preserved and/or used for downstream analysis.
This has severe constraints, which limit cultural outcomes. First, it is significantly more tedious and time-consuming than reading the protocol definition. Creating cultural collections is also tiresome and time-consuming: it takes up all of your space, consumables, and time.
This limits the throughput and, hence, the depth of the technique. In contrast, it has been demonstrated that the multiplicity of growing circumstances (nutrient availability, incubation temperature, oxidoreduction conditions, and combinations thereof) is critical for separating various libraries.11,12
Mixed colonies can arise when cells from different microbial populations, species, genera, or kingdoms coexist. Monoclonality (a culture that begins with a single clone) can be difficult, if not impossible, to accomplish via direct plating or dilution to extinction, even when streaking processes are repeated.
Third, some organisms (such as several archaea species) may be unable to grow on agar media, making streaking impossible.
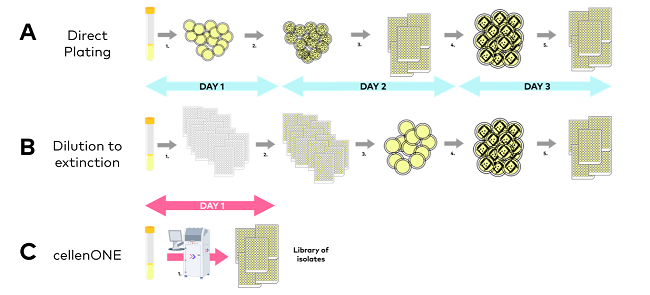
Figure 3. Classical approaches to culture library generation, i.e., Direct plating (A), and Dilution to extinction (B) vs. single microbial cell inoculation of cultures by cellenONE (C). Image Credit: Cellenion
Single-cell isolation automation with cellenONE can help alleviate all of these restrictions.
- CellenONE isolates microbial cells in various consumables, including liquids and solids, without affecting microbial development.
- Microscopy images for all separated cells can be used to check monoclonality on the instrument's screen, eliminating the need for streaking and significantly reducing experimental time (Figure 3C).
- Morphological characteristics can be used to separate cells in microbial suspensions from mixed enrichments or colonies.
- The automated device allows for high-throughput inoculation of single clones, enabling efficient one-to-many and many-to-one techniques.13
Overall, the technological innovation that enables precise single-microbial cell techniques represents a significant opportunity for all branches of microbiology.
This reflects a shift in perspective from a satellite view to a macroscopic lens, which will provide mechanistic insights into microbiota assembly and function.
References
- Prosser, J. I. (2015). Dispersing misconceptions and identifying opportunities for the use of’omics’ in soil microbial ecology. Nature Reviews Microbiology, 13(7), 439-446.
- Blainey PC. (2013). The future is now: single-cell genomics of bacteria and archaea. FEMS Microbiol Rev, 37(3), 407-27.
- Mauger, S., Monard, C., Thion, C., & Vandenkoornhuyse, P. (2022). Contribution of single-cell omics to microbial ecology. Trends in Ecology & Evolution, 37(1), 67-78.
- Persistence of free material is arguably less of an issue for metatranscriptomics and metaproteomics, as RNA and proteins have substantially shorter half-life time than DNA.
- Mauger, S., under the direction of Vandenkoornhuyse, P. & Monard, C. (2023). Elaboration of an innovative single-cell genomics approach and application to soil bacterial communities. Doctoral dissertation, Université de Rennes.
- Mauger, S., Monard, C., Thion, C., & Vandenkoornhuyse, P. (2022). Contribution of single-cell omics to microbial ecology. Trends in Ecology & Evolution, 37(1), 67-78.
- For single cell proteomics (scMS), no amplification is possible so that the feasibility of scMS applied to microbial cells rely on mass spectrometer sensitivity, which is constantly increasing these last years.
- Weber, E., under the direction of Gubry-Rangin, C. & Prosser, J. (2016). Ecological insights into unexplored Archaea through environmental ecophysiology, single-cell genomics and cultivation. Doctoral dissertation, University of Aberdeen.
- Pham, V. H., & Kim, J. (2012). Cultivation of unculturable soil bacteria. Trends in biotechnology, 30(9), 475-484.
- Kalam, S., Basu, A., Ahmad, I., Sayyed, R. Z., El-Enshasy, H. A., Dailin, D. J., & Suriani, N. L. (2020). Recent understanding of soil acidobacteria and their ecological significance: a critical review. Frontiers in Microbiology, 11, 580024.
- Lagier, J. C., Dubourg, G., Million, M., Cadoret, F., Bilen, M., Fenollar, F., … & Raoult, D. (2018). Culturing the human microbiota and culturomics. Nature Reviews Microbiology, 16(9), 540-550.
- Vanstokstraeten, R., Mackens, S., Callewaert, E., Blotwijk, S., Emmerechts, K., Crombé, F., … & Demuyser, T. (2022). Culturomics to investigate the endometrial microbiome: proof-of-concept. International Journal of Molecular Sciences, 23(20), 12212.
- For instance: one sample to many different growth conditions (media, temperature etc.), each of the many cells of a sample to wells containing a compound of interest for high throughput metabolic screening etc.
About SCIENION GmbH
SCIENION is a global market leader in ultra-low volume precision dispensing and microarray as well as biosensor technologies.
Our customers in the entire life science sector benefit from an integrated product portfolio and services advancing cost-efficient multiplex analysis, miniaturization and automation – from early research to high throughput production.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.