This article is based on a poster originally authored by David Wensel, Margaret Gartland, Jagadish Beloor, Kartika Shetty, Jana Wolf, Andrew Clark, Allan Tenorio, Max Lataillade and Mark Krystal, which was presented at ELRIG Drug Discovery 2024 in affiliation with ViiV Healthcare and Domainex Ltd.
This poster is being hosted on this website in its raw form, without modifications. It has not undergone peer review but has been reviewed to meet AZoNetwork's editorial quality standards. The information contained is for informational purposes only and should not be considered validated by independent peer assessment.

Key takeaways
- HIV-1 envelope glycoproteins with any of 7 substitutions or polymorphisms in gp120 show varying degrees of reduced susceptibility to temsavir (TMR), a small-molecule HIV-1 attachment inhibitor
- While the gp120 polymorphisms showed an altered affinity for both TMR and CD4, only TMR affinity strongly correlated with the observed reduced susceptibility to TMR, suggesting that this reduction is due to a decreased likelihood of TMR binding
- Despite the reduced TMR binding affinity observed with the gp120 polymorphisms, once TMR does bind to gp120, it retains the ability to fully block the engagement of gp120 with CD4
Introduction
- TMR, the active component of the prodrug fostemsavir, is a small-molecule HIV-1 attachment inhibitor that binds directly to the gp120 subunit within the HIV-1 envelope glycoprotein, gp160, and selectively inhibits the interaction between the virus and cellular CD4 receptors1
- Previous studies identified amino acid positions in gp120 where substitutions or polymorphisms have the potential to reduce phenotypic susceptibility to TMR2-4 (Figure 1)
- Substitutions S375H/I/M/N/Y, M426L, M434K, and M475I have been associated with higher TMR half-maximal inhibitory concentration (IC50) fold-change relative to a reference strain
- To explore the mechanism by which TMR susceptibility is altered in envelope glycoproteins with gp120 substitutions or polymorphisms, biophysical analyses were performed

Figure 1. Co-crystal Structure of Temsavir Bound to gp1205. Image Credit: Image courtesy of David Wensel et al., in partnership with ELRIG (UK) Ltd.
gp120 (green); temsavir (magenta); β20-21 loop (blue); CD4 binding loop (red).
Methods
- 7 polymorphs were engineered, either alone (S375H/I/M/N, M426L, M434I, M475I) or in combination (S375H + M475I), into the JRFL gp160 gene
- Pseudoviruses that expressed these gp160 proteins were produced and tested for sensitivity to TMR and a control compound, efavirenz
- Separately, 6xHis-tagged JRFL gp120 proteins containing these changes were expressed in 293HEK cells via a BacMam approach and purified using immobilized metal affinity chromatography
- Binding studies on the purified proteins were conducted using a Creoptix® WAVE Delta GCI system (Malvern Panalytical, Malvern, UK) to determine the impact of these polymorphs on binding to TMR and a soluble recombinant form of human CD4, as well as the ability of TMR to block gp120 binding to CD4
- Binding kinetics were determined using gp120 proteins immobilized on the chip surface via amine coupling
- Kinetic parameters that were evaluated included on-rate, off-rate, and affinity
- Competition studies were conducted with CD4 immobilized on the chip surface via amine coupling; mixtures of gp120 proteins and TMR flowed over the chip in solution
Results
Impact of gp120 polymorphisms on susceptibility to Temsavir
- The 7 individual polymorphic pseudoviruses were found to have a range of altered susceptibility to inhibition by TMR from a low of 4-fold to a high of >12,500-fold, relative to wild-type JRFL sensitivity (Table 1)
- A dual polymorph, S375H + M475I, exhibited the greatest shift in susceptibility to TMR of >29,700-fold (Table 1)
Table 1. Effect of gp120 Polymorphisms on Susceptibility to Temsavir and Efavirenz. Source: ELRIG (UK) Ltd.
Polymorphic
envelope
glycoproteina |
Temsavir |
Efavirenz |
Mean (SD)
IC50, nMb |
Mean
MPI, % |
IC50 fold
changec |
Mean (SD)
IC50, nM |
Mean
MPI, % |
IC50 fold
change |
| JRFL-WT |
0.19 (0.04) |
100.4 |
1 |
1.6 (0.4) |
100.3 |
1 |
| M426L |
86.7 (6.9) |
100.2 |
433 |
1.8 (0.4) |
100.5 |
1.1 |
| M434I |
0.8 (0.2) |
100.3 |
4 |
1.7 (0.3) |
100.4 |
1.1 |
| M475I |
14.0 (3.6) |
100.3 |
70 |
1.6 (0.4) |
100.3 |
1 |
| S375H |
178 (21) |
100.2 |
890 |
1.8 (0.4) |
100.4 |
1.1 |
| S375M |
2510 (905) |
73.6 |
12,551 |
1.6 (0.3) |
100.4 |
1 |
| S375N |
5.1 (0.5) |
100.3 |
25 |
1.7 (0.4) |
100.3 |
1.1 |
| S375I |
19.8 (1.5) |
100.4 |
99 |
1.6 (0.4) |
100.4 |
1 |
| S375H + M475I |
5945 (880) |
68.9 |
29,726 |
1.8 (0.2) |
100.6 |
1.1 |
IC50, half-maximal inhibitory concentration; MPI, maximum percent inhibition; WT, wild-type.
aPseudovirus generated to present the specified envelope glycoprotein variant (JRFL background).
bResults presented for N=3 independent experiments.
cFold-change calculated relative to JRFL-WT IC50.
Kinetics of Temsavir and CD4 binding to the 7 polymorphic gp120 proteins
- The affinity (KD) of the gp120 proteins for CD4 varied between 0.4-fold and 3-fold compared with that of wild-type JRFL gp120 (Table 2)
Table 2. Kinetic Parameters for Binding of Polymorphic gp120 Proteins to Temsavir and CD4a. Source: ELRIG (UK) Ltd.
gp120
variant |
Temsavir |
CD4 |
ka, ×104
M-1s-1 |
kd, ×10-3
s-1 |
KD,
nM |
ka' ×105
M-1s-1 |
kd, ×10-3
s-1 |
KD,
nM |
| JRFL-WT |
6.1 (1.4) |
4.6 (1.2) |
76 (4) |
3.7 (0.1) |
3.6 (0.4) |
10 (1) |
| M426L |
1.5 (0.2) |
1.2 (0.4) |
83 (34) |
1.5 (0.4) |
2.7 (0.2) |
19 (5) |
| M434I |
12.9 (0.2) |
15.2 (0.4) |
117 (4) |
1.2 (0.3) |
1.3 (0.3) |
11 (5) |
| M475I |
2.2 (0.6) |
3.4 (0.4) |
164 (42) |
1.2 (0.1) |
3.8 (0.4) |
31 (4) |
| S375H |
1.1 (0.2) |
1.2 (0.6) |
111 (44) |
3.6 (2.4) |
1.1 (0.1) |
4 (3) |
| S375M |
1.2 (0.3) |
5.6 (1.4) |
456 (36) |
2.0 (0.7) |
3.1 (0.6) |
16 (3) |
| S375N |
3.1 (0.4) |
1.7 (0.7) |
54 (22) |
1.0 (0.1) |
1.7 (0.2) |
17 (3) |
| S375I |
1.6 (0.2) |
2.8 (0.2) |
179 (19) |
2.2 (0.1) |
4.3 (0.7) |
20 (3) |
S375H +
M475I |
0.6 (0.1) |
33.5 (0.5) |
5603 (930) |
2.1 (2.0) |
0.5 (0.2) |
4 (3) |
ka, on-rate binding of temsavir or CD4 to gp120;
kd, off-rate binding of temsavir or CD4 to gp120;
KD, binding affinity of temsavir or CD4 to gp120 (KD = kd/ka); WT, wild-type.
aAll values represent mean (SD) for N=3 independent measurements.
- There was no correlation between TMR sensitivity of the individual polymorphic pseudoviruses and the on-rate, off-rate, or affinity for CD4 binding to the corresponding gp120 proteins (Figure 2)
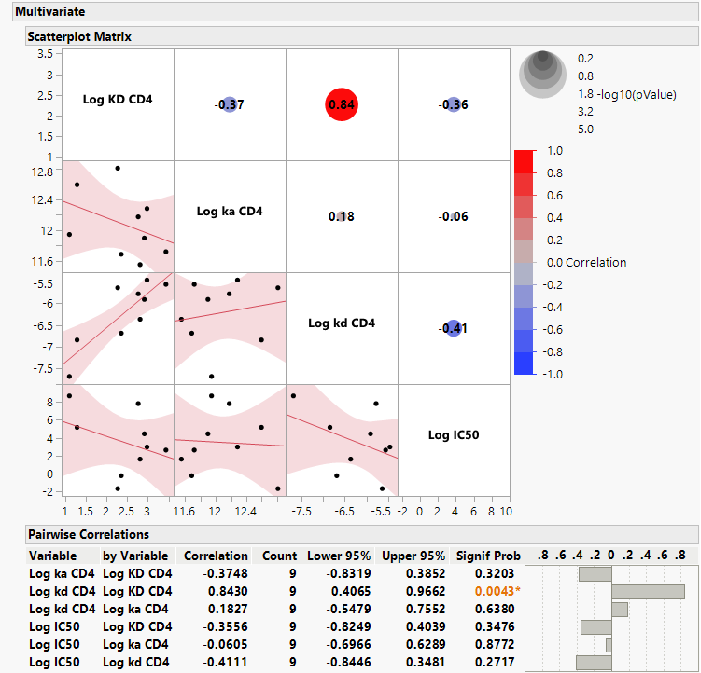
Figure 2. Lack of Correlation Between Antiviral Susceptibility and CD4 Binding Kinetics. Image Credit: Image courtesy of David Wensel et al., in partnership with ELRIG (UK) Ltd.
IC50, half-maximal inhibitory concentration; ka, on-rate of binding; kd, off-rate of binding; KD, binding affinity.
- In contrast, the affinity (KD) of TMR for the polymorphic gp120 proteins varied from 0.7-fold to 74-fold compared with wild-type JRFL gp120 (Table 2)
- A strong correlation (P=0.0011) was found between the on-rate of TMR binding to the polymorphic gp120 proteins and the susceptibility of those polymorphs to inhibition by TMR in the pseudovirus assay (Figures 3-4)
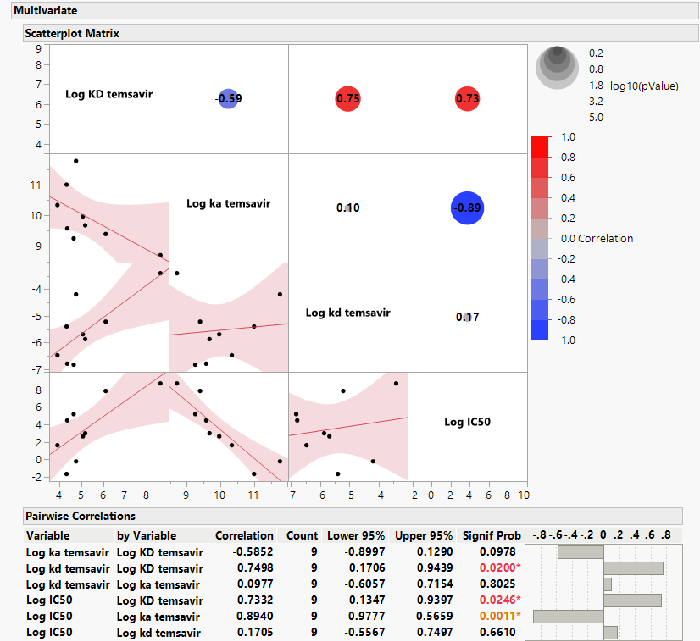
Figure 3. Correlations Between Antiviral Susceptibility and Temsavir Binding Kinetics. Image Credit: Image courtesy of David Wensel et al., in partnership with ELRIG (UK) Ltd.
IC50, half-maximal inhibitory concentration; ka, on-rate of binding; kd, off-rate of binding; KD, binding affinity.
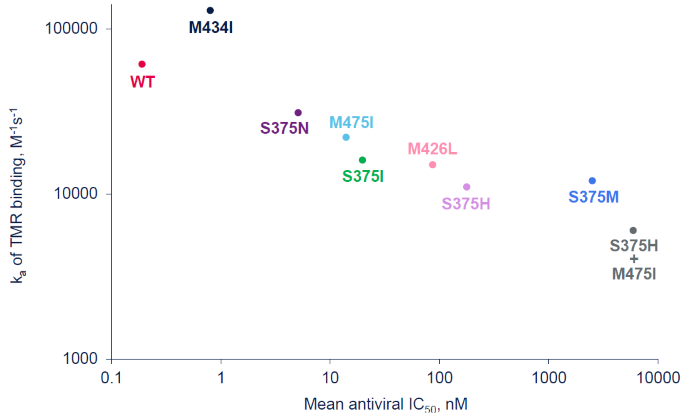
Figure 4. On-rate (ka) vs Antiviral IC50 Plot for gp120 Variants. Image Credit: Image courtesy of David Wensel et al., in partnership with ELRIG (UK) Ltd.
IC50, half-maximal inhibitory concentration; ka, on-rate of binding.
Binding competition between polymorphic gp120 proteins, CD4, and Temsavir
- In competition assays, TMR retained the capacity to fully block the binding of all polymorphic gp120 proteins to CD4, though 3 polymorphs (M426L, M475I, and S375I) required higher concentrations of TMR (Table 3)
Table 3. Degree of Inhibition of CD4 Binding by Temsavir Under Specified Conditions. Source: ELRIG (UK) Ltd.
| gp120 variant |
gp120 (2 × KD) +
temsavir (20 × KD) |
gp120 (2 × KD) +
temsavir (40 × KD) |
| JRFL-WT |
Complete |
Complete |
| M426L |
Partial |
Complete |
| M434I |
Complete |
Complete |
| M475I |
Partial |
Complete |
| S375H |
Complete |
Complete |
| S375M |
Complete |
Complete |
| S375N |
Complete |
Complete |
| S375I |
Partial |
Complete |
| S375H + M475I |
Complete |
Complete |
KD, binding affinity; WT, wild-type.
Conclusions
- The loss of susceptibility to TMR observed for these HIV-1 envelope glycoprotein polymorphisms was strongly correlated with reductions in the on-rate for TMR binding, suggesting that these polymorphic envelope proteins prefer conformations less amenable to initial TMR engagement.
- While specific polymorphisms showed an altered affinity for CD4, this did not correlate with antiviral susceptibility to TMR
- Despite reduced binding affinity, TMR could still fully block all gp120 polymorphs from binding to CD4 given sufficiently high concentrations.
- Overall, these results suggest that TMR therapy may still be of benefit to individuals with HIV strains harboring these gp120
Acknowledgments
ViiV Healthcare funded this study. Editorial assistance and graphic design support for this poster were provided under the direction of the authors by MedThink SciCom and funded by ViiV Healthcare. polymorphisms
References
- Nowicka-Sans, B., et al. (2012). In VitroAntiviral Characteristics of HIV-1 Attachment Inhibitor BMS-626529, the Active Component of the Prodrug BMS-663068. Antimicrobial Agents and Chemotherapy, 56(7), pp.3498–3507. https://doi.org/10.1128/aac.00426-12.
- Gartland, M., et al. (2022). Week 96 Genotypic and Phenotypic Results of the Fostemsavir Phase 3 BRIGHTE Study in Heavily Treatment-Experienced Adults Living with Multidrug-Resistant HIV-1. Antimicrobial Agents and Chemotherapy, 66(6). https://doi.org/10.1128/aac.01751-21.
- Lataillade, M., et al. (2018). Viral Drug Resistance Through 48 Weeks, in a Phase 2b, Randomized, Controlled Trial of the HIV-1 Attachment Inhibitor Prodrug, Fostemsavir. JAIDS Journal of Acquired Immune Deficiency Syndromes, 77(3), pp.299–307. https://doi.org/10.1097/qai.0000000000001602.
- Pancera, M., et al. (2017). Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529. Nature Chemical Biology, 13(10), pp.1115–1122. https://doi.org/10.1038/nchembio.2460.
About Domainex
Domainex is a fully integrated drug discovery CRO based near Cambridge, UK. It serves pharmaceutical, biotechnology, academic and patient foundations globally. Domainex’s drug discovery service business was established in 2001 and since that time has continued to expand to serve a wider range of international clients including UCB, FORMA Therapeutics, St George’s University, and The Institute of Cancer Research.
About ELRIG (UK) Ltd.
The European Laboratory Research & Innovation Group (ELRIG) is a leading European not-for-profit organization that exists to provide outstanding scientific content to the life science community. The foundation of the organization is based on the use and application of automation, robotics and instrumentation in life science laboratories, but over time, we have evolved to respond to the needs of biopharma by developing scientific programmes that focus on cutting-edge research areas that have the potential to revolutionize drug discovery.
Comprised of a global community of over 12,000 life science professionals, participating in our events, whether it be at one of our scientific conferences or one of our networking meetings, will enable any of our community to exchange information, within disciplines and across academic and biopharmaceutical organizations, on an open access basis, as all our events are free-of-charge to attend!
Our values
Our values are to always ensure the highest quality of content and that content will be made readily accessible to all, and that we will always be an inclusive organization, serving a diverse scientific network. In addition, ELRIG will always be a volunteer led organization, run by and for the life sciences community, on a not-for-profit basis.
Our purpose
ELRIG is a company whose purpose is to bring the life science and drug discovery communities together to learn, share, connect, innovate and collaborate, on an open access basis. We achieve this through the provision of world class conferences, networking events, webinars and digital content.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.
Last Updated: Nov 18, 2024