The history of DNA started as early as 5000 BC, when humans began the practice of selective breeding to produce more robust crops and livestock. There have been many instances throughout history where notable discoveries have been made about DNA and inheritance; these have formed the foundations of what we know and continue to advance today. This article aims to provide a brief overview of the history of DNA research, leading up to Watson and Crick's discovery of the structure of DNA, and what came next.
 HelloRF Zcool | Shutterstock
HelloRF Zcool | Shutterstock
Skip to:
Greek philosophers explored the idea of human inheritance some 1600 years after 5000 BC. The notable Aristotle suggested that traits acquired throughout an organism’s lifetime could be transmitted to their offspring. Aristotle supplemented this hypothesis with the theory of ‘pangenesis’ which described how these traits could be passed on; particles called ‘gemmules’ encapsulated these traits and allowed them to be transmitted to reproductive cells.
The first instance of active research, however, began with Augustinian monk Gregor Mendel. He is commonly known today as the ‘Father of Genetics’. His discoveries marked the advent of modern genetics.
Gregor Mendel: The 'Father of Genetics’
Gregor Mendel was a monk who performed a meticulous series of experiments with pea plants in 1857. Mendel selected specific characteristics of pea plants to study. For each characteristic (or phenotype), he obtained lines of plants that were pure, producing offspring with characteristics identical to the parent.
Mendel then used these pure-breeding plants in subsequent experiments to perform reciprocal crosses – breeding plants of different characteristics and examining their progeny. He repeated this over two generations of plants and found that he could obtain consistent ratios of traits. This allowed him to suggest theories to explain these ratios. In turn, he deduced four important principles of inheritance;
- Hereditary determinants are called genes.
- Genes exist in pair, called alleles which may be dominant or recessive.
- Genes are segregated in the gametes which are consequently carriers of only one gene pair.
- Fertilization, in which two gametes fuse, is random.
Friedrich Miescher and Richard Altmann
Swiss physician Friedrich Miescher discovered a substance he called ‘nuclein’ in 1869. To do so, he devised a protocol to isolate DNA. Later, he isolated a comparatively purer sample of this same material from the sperm of salmon. This formed the basis for his paper in 1871. In 1889, his pupil, Richard Altmann renamed nuclein to ‘nucleic acid’. This substance was found to exist only in the chromosomes. This discovery built on earlier work by Walter Flemming who described the appearance and behavior of chromosomes in 1882.
Theodor Boveri and Walter Sutton
In 1902, Theodor Boveri and Walter Sutton independently postulated that chromosomes were not only the carriers of hereditary units but were organized so that different locations of the chromosomes corresponded to specific hereditary traits. Boveri did this by examining chromosomal behavior during cell division and gamete formation, and later, Sutton emphasized how this was congruent with Medel’s findings. This formed the basis of cytogenetics which describes the structure, function and inheritance of chromosomes.
Frederick Griffith
Frederick Griffith was a scientist was working on a project in 1928 that formed the basis that DNA was the molecule of inheritance. Griffith's experiment involved mice and two types of pneumonia – one was virulent and characterizable by a rough appearance, and the other non-virulent, and visually distinguishable by a smooth coat. Injection of virulent pneumonia into a mouse caused the mouse to die.
The opposite response occurred when non-virulent pneumonia was injected into a mouse; this mouse survived. However, he then observed that mixing a dead virulent strain with a living non-virulent bacterial strain enabled the former to be transformed into the latter. When injected into a mouse, the mouse was killed.
When he isolated the bacteria of the dead mouse, he noticed that they now possessed a smooth capsule characteristic. This led him to hypothesize that the previously rough non-virulent strain had acquired some substance from the virulent strain. He termed this substance the ‘transforming principle’, which he believed to be the inheritance molecule. This passing on of the inheritance molecule was what he called transformation.
Phoebus Levene
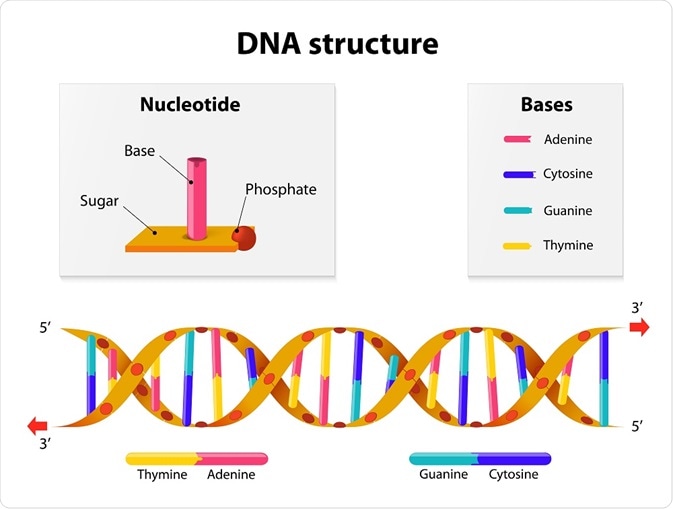
In 1929 Phoebus Levene at the Rockefeller Institute identified the components that make up a DNA Molecule. Those components are:
- The four bases
- Adenine (A)
- Cytosine (C)
- Guanine (G)
- Thymine (T)
- Sugar
- Phosphate
He showed that the components of DNA were linked in the order phosphate-sugar-base. Crucially, he distinguished the two ribose subtypes – deoxyribose and ribose. Levene coined the arrangement of the sugar, base and phosphate group a ‘nucleotide’.
However, Levene thought the chain was short and that the bases repeated in the same fixed order. It was Torbjorn Caspersson and Einar Hammersten who showed that DNA was, in fact, a polymer.
Oswald Avery
Oswald Avery continued with Griffith’s experiment, and accumulated evidence to suggest that the integrity of the capsule was essential for virulence. He employed a novel method of transforming non-virulent bacteria into encapsulated virulent bacteria, performing the transformation in culture, rather than in mice. Avery and his colleagues used a process of elimination to determine the identity of the ‘transforming principle’.
Avery used hydrolytic enzymes that targeted either protein, DNA or RNA respectively. Following these respective treatments, these treated extracts were mixed with rough, nonvirulent strains. They found that transformation of these strains occurred with all treated extracts, except those in which the extract was mixed with DNase. This provided evidence that DNA, and not any other component, was the ‘transforming principle’. He had found the basis of the inheritance.
Colette and Roger Vendrely and André Boivin
In 1949 André Boivin and his students Colette and Roger Vendrely found that the nuclei of germ cells contained only half the amount of DNA than that of somatic cells. This demonstrated that the genetic content was consistent through all cells in the body, and within a member of the same species. The basis of this constancy came from the process of gametogenesis. This provided yet more evidence that DNA was the ‘transforming principle’.
Erwin Chargaff and Chargaff’s rule
In the 1940s Erwin Chargaff found that the base composition (adenine, guanine, cytosine, and thymine) differed between species and that ratios between them were invariable; the quantity of adenine was equal to that of thymine. The same ratio of 1:1 was seen for cytosine and guanine. This discovery later became known as Chargaff’s Rule.
Rosalind Franklin
In 1952, British researcher Rosalind Franklin crystallized a molecule of DNA. From the X-ray diffraction images Franklin obtained, she demonstrated that DNA contained a regularly repeating helical structure. The images allowed precise calculations of the molecular spacing in DNA
Watson and Crick
Building on Franklin’s work two scientists, James Watson and Francis Crick, made a model of the DNA structure approximately 2 years later.
Their model was that of a double helix that consisted of evenly spaced pairs of bases connecting the two strands. It was possible to predict the measurements between bases and the number of bases per turn; further, there were strict base-pairing rules. To account for their measurements, they discovered that Thymine could only pair with Adenine and Guanine with Cytosine. This concurred with Chargaff's rule.
 Designua | Shutterstock
Designua | Shutterstock
Notable discoveries beyond the double helix
A decade later, Robert W. Holley, Har Gobind Khorana and Marshall W. Nirenberg won a Nobel prize for their work deciphering how DNA related to protein synthesis. They established the dogma of information transfer from DNA to RNA to protein.
In 1977, Frederick Sanger, Allan Maxam, and Walter Gilbert developed methods to sequence DNA. This was supplemented in 1983 by Kary Mullis, who invented the polymerase chain reaction (PCR) to amplify DNA. Together these methods paved the way for sequencing of the human genome which began in 1990 and was completed 13 years later, in full.
Today, the focus on DNA has exploded to include ways of ‘editing’ the genome, using novel methods to change the information it encodes in highly specific ways. Further, lesser known areas of the genome are under study; the field of epigenomics is rapidly expanding, allowing us to understand how and why genome behavior differs substantially between individuals.
Further Reading
Last Updated: May 1, 2019