Restriction fragment length polymorphism (RFLP) is a technique invented in 1984 by the English scientist Alec Jeffreys during research into hereditary diseases. It is used for the analysis of unique patterns in DNA fragments in order to genetically differentiate between organisms – these patterns are called Variable Number of Tandem Repeats (VNTRs).
Genetic polymorphism is defined as the inherited genetic differences among individuals in over 1% of normal population. The RFLP technique exploits these differences in DNA sequences to recognize and study both intraspecies and interspecies variation.
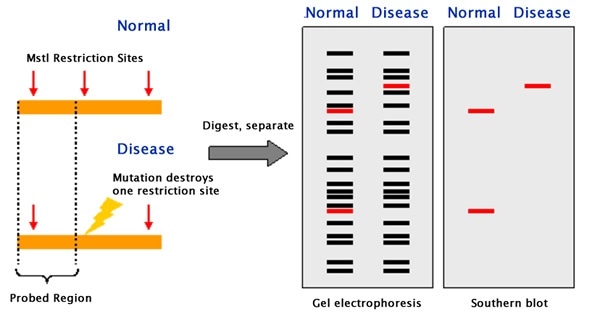
Restriction Fragment Length Polymorphism (RFLP)
Principle
Restriction endonucleases are enzymes that cut lengthy DNA into short pieces. Each restriction endonuclease targets different nucleotide sequences in a DNA strand and therefore cuts at different sites.
The distance between the cleavage sites of a certain restriction endonuclease differs between individuals. Hence, the length of the DNA fragments produced by a restriction endonuclease will differ across both individual organisms and species.
How does it Work?
RFLP is performed using a series of steps briefly outlined below:
DNA Extraction
To begin with, DNA is extracted from blood, saliva or other samples and purified.
DNA Fragmentation
The purified DNA is digested using restriction endonucleases. The recognition sites of these enzymes are generally 4 to 6 base pairs in length. The shorter the sequence recognized, the greater the number of fragments generated from digestion.
For example, if there is a short sequence of GAGC that occurs repeatedly in a sample of DNA. The restriction endonuclease that recognizes the GAGC sequence cuts the DNA at every repetition of the GAGC pattern.
If one sample repeats the GAGC sequence 4 times whilst another sample repeats it 2 times, the length of the fragments generated by the enzyme for the two samples will be different.
Gel Electrophoresis
The restriction fragments produced during DNA fragmentation are analyzed using gel electrophoresis.
The fragments are negatively charged and can be easily separated by electrophoresis, which separates molecules based on their size and charge. The fragmented DNA samples are placed in the chamber containing the electrophoretic gel and two electrodes.
When an electric field is applied, the fragments migrate towards the positive electrode. Smaller fragments move faster through the gel leaving the larger ones behind and thus the DNA samples are separated into distinct bands on the gel.
Visualization of Bands
The gel is treated with luminescent dyes in order to make the DNA bands visible.
Applications of RFLP
RFLP has been used for several genetic analysis applications since its invention.
Some of these key applications of RFLP are listed below:
- To determine the status of genetic diseases such as Cystic Fibrosis in an individual.
- To determine or confirm the source of a DNA sample such as in paternity tests or criminal investigations.
- In genetic mapping to determine recombination rates that show the genetic distance between the loci.
- To identify a carrier of a disease-causing mutation in a family.
Disadvantages of RFLP
Since its invention, RFLP has been a widely used genome analysis techniques employed in forensic science, medicine, and genetic studies. However, it has become almost obsolete with the advent of relatively simple and less expensive DNA profiling technologies such as the polymerase chain reaction (PCR).
The RFLP procedure requires numerous steps and takes weeks to yield results, while techniques such as PCR can amplify target DNA sequences in a mere few hours.
Additionally, RFLP requires a large DNA sample, the isolation of which can be a laborious and time-consuming process. In contrast, PCR can amplify minute amounts of DNA in a matter of hours.
Due to numerous reasons such as these, the PCR technique has largely replaced RFLP in most applications requiring DNA sequencing such as paternity testing or forensic sample analysis.
Furthermore, the identification of single-nucleotide polymorphisms in the Human Genome Project has almost replaced the need for RFLP in disease status analysis.
References
Further Reading
Last Updated: Jun 26, 2019