The advent of fluorescence microscopy has made it possible to visualize proteins inside live cells. This is particularly useful when studying the location of signaling pathways and binding partners.
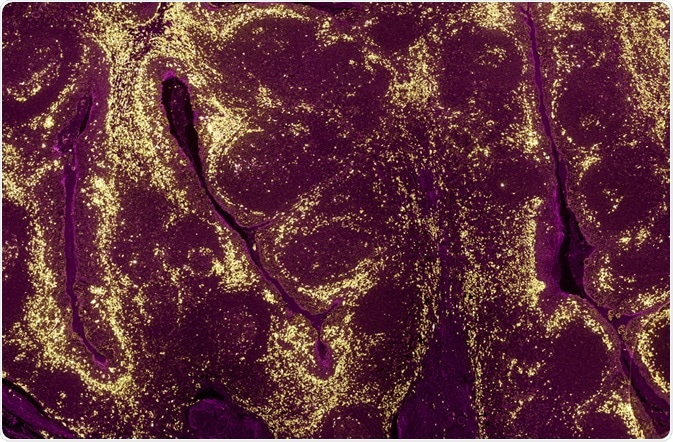
A tumour sample where the levels of different proteins in cell populations have been identified using a fluorescently labelled antibody and fluorescence microscopy. (Carl Dupont | Shutterstock)
Studying protein localization
The function of a protein can be inferred by its localization, particularly when a signaling pathway is known. The location of the protein can be determined through the use of recombinant proteins fused with a reporter molecule, fluorescent dyes, or protein-specific fluorescent antibodies.
Fluorescent proteins recombined with either target protein or regulatory regions can be overexpressed inside cells to understand fundamental cellular processes. An advantage of using genetic systems to express proteins is that the fluorescent proteins can be selectively activated in certain regions of tissues or cells. This, in turn, makes the process of visualizing proteins easy.
Studying protein structure
The methods previously employed to study protein structure include cryo-electron microscopy, protein NMR, and X-ray crystallography. In cryo-electron microscopy, high energy electrons are directed at the sample to form a 2D projection. After capturing the 2D orientation of molecules from all angles, a 3D image is reconstructed. However, this process cannot be performed in live samples and it requires extensive sample preparation.
On the other hand, nuclear magnetic resonance (NMR) captures the magnetic and chemical properties of the atoms but requires a pure solution of the sample which is again not optimal for studying cells. X-ray crystallography requires the sample to be crystallized before it can be imaged. In this case, the process of crystallization can alter the native conformation of proteins.
In contrast, super-resolution methods, such as photoactivated localization microscopy (PALM) or stimulated emission depletion microscopy (STED) have a very high resolution of 20-30 nanometers. This is below the diffraction limit of conventional microscopes (250 nanometers). Using these methods, protein and membrane structure can be visualized to a great detail in their in vivo settings.
Studying protein dynamics
Using a method called fluorescent recovery after photobleaching or FRAP, it can be measured how a protein moves or diffuses inside a cell. The diffusion of a protein is related to its folded or misfolded status, its stability, and binding to other molecules. Thus, understanding its motion or diffusion can inform as to its structure and function inside cells.
In this method, the fluorescent molecules in a specific region of a cell are completely bleached by shining an intense and constant beam of light. Then, the recovery of the fluorescence in the bleached region is measured as a function of time to map its diffusion parameters.
Studying protein signaling and interaction
This is determined using a fluorescence method called Förster resonance energy transfer or FRET. In this method, there is a transfer of energy between two closely placed molecules conveniently called donor and acceptor molecules.
The donor chromophore is initially in an excited state and as it is close in proximity to another molecule, it transfers its energy to a donor chromophore. This transfer is, thus, proportional to the sixth power of the distance between donor and acceptor.
Hence, based on the efficiency of the transfer of energy, the distance between two molecules or protein domains can be inferred. This allows scientists to determine which protein domains are interacting during a signaling event.
Studying protein levels
Apart from signaling, structure, and function, the fluorescence of a protein can also be used to measure the levels of a protein expressed inside a cell or a tissue. This can be used in cases where the fluorescent probe is recombined with the gene or reporter of a gene. The expression of the reporter, in this case, reflects the levels of protein it is recombined with.
Further Reading
Last Updated: Nov 28, 2018