Mar 1 2005
Each year, there are an estimated 50 million cases of amoebic dysentery, causing up to 100,000 deaths, mostly in developing countries. On Thursday 24 February 2005, researchers at the Wellcome Trust Sanger Institute and their colleagues reported in Nature magazine the genome sequence of the parasite that causes the disease, Entamoeba histolytica.
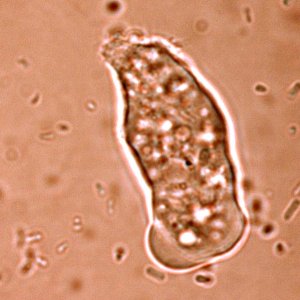
A striking new picture emerges of an organism adapting itself to life as a parasite of the human gut. It seems to have jettisoned many genes required for independent survival, relying instead on us, its human host, to help it out. At the same time, it appears to have picked up genes from bacteria in its environment - genes that allow it to use a wider range of sugars and other energy sources.
Dr Matt Berriman, Project Leader at the Wellcome Trust Sanger Institute, said, "The results give a fascinating glimpse of how this ancient parasite evolved and highlights unusual metabolic processes that may be exploitable as drug targets. The genome is full of enzymes and transporters that could be followed up by experimentation."
E. histolytica is a remarkable organism. Its name derives from the fact that it "eats" tissues (histo-, tissue; lysis, loosen): it also has a voracious appetite for bacteria, which is how it can be grown in test tubes. Amoebic dysentery is caught by ingesting cysts - dormant organisms - from water, food or hands contaminated with human faeces. Inside the gut of human host, the parasite hatches and begins multiplying.
The sequencing of E. histolytica was a collaborative effort led by the Wellcome Trust Sanger Institute and The Institute for Genomic Research (TIGR) in Rockville, MD, USA. The project was supported by grants from the Wellcome Trust and from the National Institute of Allergy and Infectious Diseases (NIAID), which is part of the National Institutes of Health.
The sequence reveals almost 10,000 genes, some of which appear to have been acquired from bacteria. Nearly one-third of the genes are unrelated to genes in public databases.
"Entamoeba histolytica is a major cause of acute diarrhoea in developing countries, but the research community working to combat it is relatively small," continued Dr Berriman. "The draft sequence provides a much-needed boost, and our continuing efforts to improve our understanding will strengthen these efforts."
A defining feature of E. histolytica is its voracious appetite - it lives in an environment where it is surrounded by and eats bacteria. In the human gut, it causes diarrhoea, fluid and salt loss and bleeding from the gut. It can also invade the gut wall and spread to other tissues, causing more serious disease.
"Clearly, this amoeba has genes that allow it to sense certain facets of its environment and respond to those cues," said Neil Hall, a Sanger Institute scientist, now at TIGR, who is the senior author of the Nature study.
It was long considered to be a "primitive" organism - originating from the time that bacteria and more complex organisms diverged in evolution, because it lacks many of the characteristic components of more complex cells. It also has some features typical of bacteria.
One emerging and strengthening picture from the sequence of parasite genomes is the number of genes that successful organisms use to evade our defence mechanisms. E. histolytica has large "families" of genes used to produce its surface coat. It is thought that it may use different coats to prevent detection in the human body for years at a time.
The range of genetic tricks surprised the research team, but also leads to hope. "It is also unusual for so many bacterial-like genes to be used in this way," commented Dr Berriman. "Taken together these findings open up a lot of possibilities for producing selective inhibitors - drugs that affect the organism but don't affect us."
http://www.sanger.ac.uk