A cutting-edge genomic model reveals where your Salmonella risk really lies—spotlighting chicken and veggies as major sources and reshaping how we tackle foodborne illness.
 Study: Attribution of Salmonella enterica to Food Sources by Using Whole-Genome Sequencing Data. Image Credit: nobeastsofierce / Shutterstock
Study: Attribution of Salmonella enterica to Food Sources by Using Whole-Genome Sequencing Data. Image Credit: nobeastsofierce / Shutterstock
In a recent study published in the journal Emerging Infectious Diseases, a group of researchers used genome sequencing and machine learning to determine the primary food sources causing human Salmonella infections in the United States (US).
Background
Every year, Salmonella enterica infections result in approximately 1.35 million illnesses, leading to significant hospitalizations in the US. Common sources include contaminated food, water, animals, soil, and infected individuals. Serotypes such as Enteritidis and Typhimurium can infect numerous hosts, whereas others like Dublin primarily affect cattle. Traditional methods attribute only about 5% of cases to known outbreaks, leaving most illnesses untracked. Previous approaches relied on limited laboratory techniques, but with the adoption of Whole-Genome Sequencing (WGS), a clearer picture of Salmonella transmission pathways can emerge. Enhanced attribution models are critical to refining food safety regulations and preventive actions, emphasizing the need for continued research using advanced genomic technologies.
About the study
Researchers compiled a dataset of 18,661 Salmonella isolates sourced from food and animal samples available in the National Center for Biotechnology Information (NCBI), augmented by metadata from US governmental agencies, including the Food and Drug Administration (FDA), United States Department of Agriculture’s Food Safety and Inspection Service (USDA-FSIS), and Centers for Disease Control and Prevention (CDC). Isolates were categorized into 15 distinct food groups, excluding mixed-source samples. Due to an excess of chicken isolates, 50% were randomly selected to balance the dataset, and inverse class weighting was applied to further correct imbalances. Although the model used global Salmonella isolates, 76% were from the United States, making it broadly representative of domestic food sources.
For human infections, 6,470 Salmonella isolates with unknown infection sources and no international travel history were collected from the Foodborne Diseases Active Surveillance Network (FoodNet), covering about 15% of the US population between 2014 and 2017.
The research team assembled genetic data using SPAdes software and applied whole-genome multilocus sequence typing (wgMLST) to characterize both food-derived and human isolates. Serotype identification utilized the SeqSero2 tool. A Random Forest machine learning algorithm, which classifies data using numerous genetic markers, was trained on isolates with known sources. The model was evaluated for accuracy using cross-validation and permutation importance to identify the most informative genomic markers. The model achieved maximum accuracy using a subset of 7,360 genetic loci, reinforcing the value of high-dimensional genomic data for classification tasks. The optimized model predicted infection sources for human cases with >50% probability, attributing uncertain cases to unknown sources.
Study results
The Random Forest model, trained on genomic data from 18,661 food and animal-derived isolates, identified chicken (31%), vegetables (13%), turkey (12%), and pork (11%) as the predominant Salmonella sources. The most prevalent Salmonella serotypes were Kentucky, Typhimurium, Enteritidis, and Heidelberg.
Applied to human infections, the model analyzed 6,470 cases and attributed 34% of illnesses to chicken and 30% to vegetables, accounting for nearly two-thirds of infections. When uncertainty was considered (probabilities <50%), about 44% of cases remained unclassified. Excluding uncertain cases, the model attributed 46% of infections to chicken and 27% to vegetables, collectively accounting for roughly 73% of confirmed sources.
Different Salmonella serotypes showed distinct source associations. Chicken was notably linked to serotypes Enteritidis, Typhimurium, Heidelberg, and Infantis, while vegetables were primarily associated with Javiana and Newport. Pork emerged as the dominant source for serotype Salmonella enterica 4,[5],12:i:− (STM).
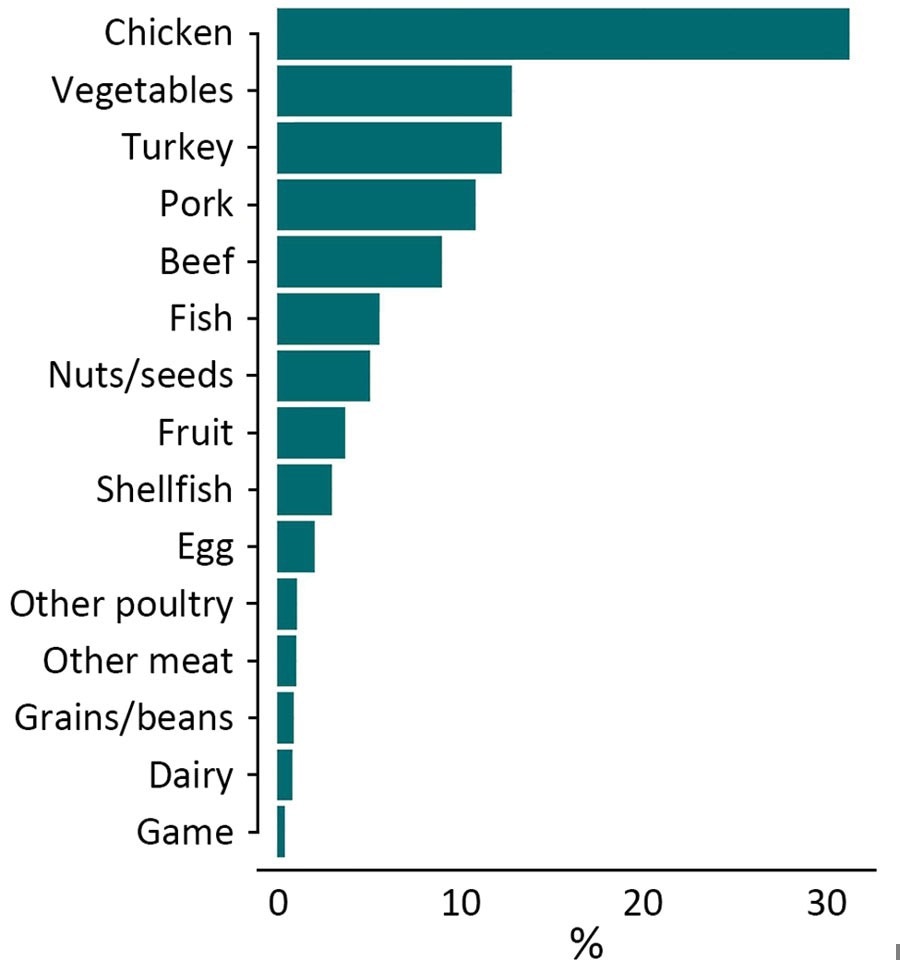
Percentage of Salmonella isolates collected from known single source foods in the United States and other countries from 2003–2018 (used as training data in random forest model), by food category (N = 18,661, including 613 isolates collected before 2003).
The model's accuracy was strong, particularly in identifying chicken (97% accuracy), vegetables (82%), turkey (88%), pork (83%), and beef (77%). However, it struggled with less common sources like dairy and game. Increasing the number of genomic loci used improved accuracy, confirming the effectiveness of WGS and machine learning for source attribution.
Compared to previous outbreak-focused studies, this analysis highlighted chicken as a far more substantial source of Salmonella infections, reflecting different risk profiles between sporadic infections and outbreaks. Importantly, predictions aligned well with known epidemiological data, affirming the model’s real-world applicability.
These findings underscore the need for targeted interventions and policies focusing on poultry and fresh produce, which is critical for reducing the Salmonella burden in public health settings. Given that many infections remain unattributed, expanding the dataset with more diverse non-chicken isolates and additional non-food sources like environmental and wildlife samples could further enhance accuracy. The regional limitations of FoodNet data and variations in healthcare-seeking behavior also suggest the necessity for broader, nationwide data collection.
Conclusions
To summarize, this study demonstrated the effectiveness of WGS combined with a Random Forest machine learning algorithm to accurately identify food sources of Salmonella infections in the US. Chicken and vegetables emerged as primary contributors, reinforcing the importance of targeted regulatory and public health strategies. This genomic approach offers significant improvements over traditional methods, providing detailed insights crucial for food safety policy, routine surveillance, and outbreak management. Continued research should integrate broader sample diversity, expand geographic representation, and include non-food sources to further strengthen the model’s precision, benefiting public health efforts against Salmonella.