Apr 26 2011
Analysis of mutations of the 2009 pandemic influenza A(H1N1) virus by researchers at the RIKEN Omics Science Center (OSC) has revealed major genetic differences between the virus in its early phase of infection in Japan and in its peak phase. While yielding valuable clues on the genetic origins of drug resistance, the findings also pave the way toward the development of new diagnostic kits for detecting and preventing the spread of global pandemic diseases.
A unique triple combination of bird, swine and human flu viruses, the pandemic influenza A(H1N1) virus, first detected in April of 2009, quickly spread from Mexico to locations across the world. By April 2010, outbreaks of the disease at both local and global scales had resulted in roughly 18,000 deaths worldwide, causing serious damage both to human health and on the global economy.
In Japan, the first case of the pandemic was reported on May 9, 2009, thereafter spreading to hundreds of people in Osaka and Kobe and eventually leading to more than 200 deaths in the country. Existing research on the spread of the virus in Japan has provided valuable information on local strains during the early phase of infection and on their classification into different groups. How the pandemic evolved to reach its peak phase of contagion, however, is not yet well understood.
To clarify the genetic basis for this evolution, the OSC group studied 253 samples of the virus collected from the Osaka area during the initial phase (May, 2009) and from the Kansai and Kanto areas during the peak phase (October, 2009 to January 2010) of contagion. Of 20 different mutation groups identified in the peak infection group, analysis revealed that 12 were entirely new to Japan. Rapid mutation of the virus strains was traced to a genome with an extremely high evolutionary rate.
Among the variety of mutants discovered, the researchers were able to pinpoint two mutations which clearly differentiate the early phase and peak phase viruses. They also identified mutations in some viruses which confer resistance to Oseltamivir (Tamiflu), one of the most widely-used antiviral drugs. Published in the journal PLoS ONE, the findings together mark a major advance in efforts to understand the genetic origins of the 2009 A(H1N1) virus, and a key step in OSC-centered efforts to develop on-site detection techniques for controlling infection of deadly pandemics.
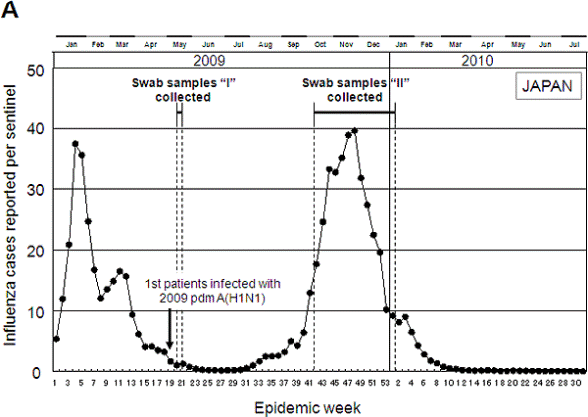
Collection of swab samples during initial and pandemic periods in Japan.
(A) The number of influenza cases reported per sentinel provider in Japan and the collection of swab samples from outpatients in Osaka, Tokyo, and Chiba areas. Swab samples were collected during initial and pandemic periods that are indicated by "I" (May 16 to May 20, 2009) and "II" (October 13, 2009 to January 6, 2010), respectively. In Japan, the first patients (three Japanese men) infected with the 2009 pdm A(H1N1) were discovered on May 9 (week 19), 2009.
Collection of swab samples during initial and pandemic periods in Japan.
(B) Mapping of the hospitals, clinics, and public health institutes that contributed to our collection of swab samples from influenza patients.
- For more information, please contact
-
- Dr. Toshihisa Ishikawa
- LSA Technology Development Unit
- RIKEN Omics Science Center
- Tel: +81-(0)45-503-9222 / Fax: +81-(0)45-503-9216
- Ms. Tomoko Ikawa (PI officer)
- Global Relations Office
- RIKEN
- Tel: +81-(0)48-462-1225 / Fax: +81-(0)48-463-3687
- Mail: [email protected]