The biological mesoscale range includes biological structures that range from 10 to 100 nanometers (billionths of a meter).
Structures in this size range include viruses, cellular organelles, large molecular complexes, and any other internal cellular environments within that range.
The cell is a hierarchy of structures that span from atoms to organelles, all of which interact in an intricate choreography with tempos that range from femtoseconds to hours.
The mesoscale is important because it represents the range of scales of cellular systems that are not fully accessible to a single experimental technique.
Why has it been difficult to visualise structures in this range, when scientists have been able to study both larger and smaller biological structures, such as cells and proteins respectively?
The real problem is not visualizing isolated component structures but rather visualizing the complex molecular environment of these component structures as they exist in a living cell.
Atomic level structures of large molecular assemblies such as the ribosome are within this mesoscale range and have been visualized with x-ray crystallography, but in isolation.
Larger cellular structures such as organelles are also within this range and can be visualized with electron or even light microscopy, but not at the atomic level.
Unfortunately there does not exist a single experimental technique that can visualize the 3 dimensional structure of cellular environments at the molecular scale.
How does the cellPACK software overcome these issues?
The cellPACK software uses structural data for a given mesoscale environment gathered from different experimental methods, which can include atomic level structures of individual biological molecules (proteins, DNA, RNA, etc.), electron microscopic structures of isolated molecular assemblies and the shapes and locations of cellular organelles, and light microscopy that can show locations of fluorescently tagged molecules in cellular environments, as well as genomic and proteomic data that describe the expression levels of the molecular components and their relative abundance or concentrations in cellular environments.
Then cellPACK takes all of this information and synthesizes one or more 3D models that are statistically consistent with all of this available information.
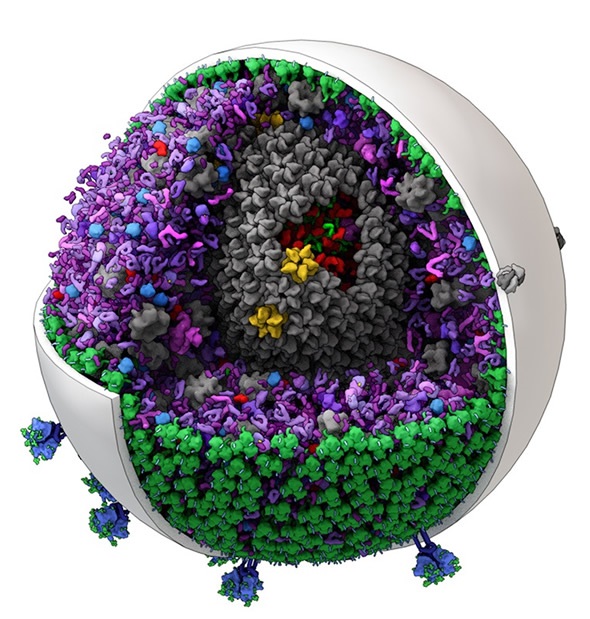
The new software can generate editable models of mid-size biological structures such as this one of HIV. (Image created by Graham Johnson and Ludovic Autin of The Scripps Research Institute.)
For a given cellular or subcellular structure, the geometry of the large components such as organelles or intact virions seen with electron microscopy can define specific volumes and surfaces to fill with the smaller molecular components.
Since the locations of the contents of these environments are constantly changing, cellPACK uses statistical measures to place these molecular components into the compartmental volumes and membrane surfaces. Thus a filled model is one snapshot of many possible fills.
Please can you outline how the cellPACK software was developed?
The cellPACK project was initiated by Graham Johnson, who came to my lab to earn a Ph.D. in structural biology. He had and art background and had worked as a scientific and medical illustrator for over 10 years.
He was inspired by the work of David Goodsell in my lab. David is a great scientist as well as a talented artist, and is well known for his watercolors of crowded molecular environments which are produced from extensive research collating the types of structural information described above.
Graham’s goal was to transform David’s illustrations into computer generated 3D models. We recognized that to do this we would need to develop a computational framework that would enable the packing of complex shapes at relatively high densities into defined volumes and onto defined surfaces.
After a number of different attempts and with the coding capabilities of Ludovic Autin, a postdoctoral researcher in my lab, the autoPACK framework was developed, which uses a “smart grid” approach to enable successive statistical placement of lists of differently shaped objects with into the volumes with without overlaps.
The cellPACK component was developed to provide the biological information that enables different types of filling algorithms for different biological components, such as globular proteins and fibrous assemblies such as DNA or RNA.
Currently, what models has the cellPACK software been used to create?
We have created molecular models of blood serum, the intact HIV virion, and packing of synaptic vesicles.
We also have a preliminary model of a mycoplasma, one of the smallest complete prokaryotic cells.
How efficient is this software to use in comparison to previous models made by hand?
cellPACK is many thousands of times more efficient than creating the models by hand. Several years ago it took me several weeks to create a model of small volume of blood plasma. Given the same starting information, cellPACK can create that model in seconds.
I believe that creating such models for simulation purposes has been a significant bottleneck in the field of large system dynamic simulations. cellPACK can remove that bottleneck and enable multiple simultaneous simulations using multiple statistically valid models of the same system.
The cellPACK software has been made free and open source. What do you hope this will achieve?
We see cellPACK as a community tool. There is an ever-increasing amount of structural data on different cell types and their components. The models that we build today will change as more data becomes available. This requires input from scientists around the world.
We hope that cellPACK will provide a framework for creating ever-improving models of cellular environments. We also hope that other developers will improve and expand cellPACK’s capabilities to represent the large variety of structures and structural hierarchies that exist in cells.
How many people have downloaded the software so far and what feedback have you received?
So far we have had about 8000 downloads of the cellPACK software. Most of these downloads originated from a crowdsourcing challenge that Graham’s and my lab with the help of AutoDesk put out to the public.
We received over 100 entries to the challenge mostly from the computer graphics and illustration community, which produced some amazing high quality images and videos of the HIV virion in blood serum (see https://cgsociety.org/).
What impact do you think the cellPACK software will have on structural biology?
In the short term we think that cellPACK will enable the community to create and share structural models of complex molecular environments and enable new analyses and simulations of these environments.
In the longer term, it is our hope that cellPACK will serve as a catalyst to bring structural biology, systems biology and synthetic biology together into a new discipline.
To this point these have been three isolated, but powerful approaches. We believe that understanding the structural mechanisms of cells at the molecular level will inform how biology works at a systems level, and enable the synthesis of new synthetic biology constructs
Where can readers find more information?
Readers can go to the cellPACK website: http://cellpack.org.
There are now two papers published on the work:
3D molecular models of whole HIV-1 virions generated with cellPACK
Graham T. Johnson,*abc David S. Goodsell,c Ludovic Autin,c Stefano Forli,c Michel F. Sanner and Arthur J. Olson, Faraday Discuss., 2014,169, 23-44
cellPACK: a virtual mesoscope to model and visualize structural systems biology
Graham T Johnson, Ludovic Autin, Mostafa Al-Alusi, David S Goodsell, Michel F Sanner & Arthur J Olson, Nature Methods (2014) doi:10.1038/nmeth.3204
About Professor Art Olson
 Arthur Olson is the Anderson Research Chair Professor in the Department of Integrative Structural and Computational Biology at The Scripps Research Institute and founder and director of its Molecular Graphics Laboratory.
Arthur Olson is the Anderson Research Chair Professor in the Department of Integrative Structural and Computational Biology at The Scripps Research Institute and founder and director of its Molecular Graphics Laboratory.
He received his Ph.D. in Physical Chemistry from the University of California, Berkeley. He was a Damon Runyon Postdoctoral Fellow at Harvard University, working on the first atomic resolution structure of an intact viral capsid.
He is currently Director of an NIH Center on the Structural Biology of AIDS which brings together 7 institutions around the U.S. to understand and develop new therapies to counter the development of drug resistance in HIV infected patients.
Olson is a pioneer in the analysis and visualization of biological assemblies spanning length scales from microns to angstroms. His laboratory has developed, applied and distributed a broad range of molecular modeling and visualization software tools over the past 30 years, including AutoDock, which is the worlds most highly cited ligand protein docking code.
In 2000 he started the first Internet distributed biomedical computing project, FightAIDS@Home, which is now running on almost 3 million computers worldwide, and which was honored by resolution in the California State Legislature.
His current work in molecular graphics focuses on the development of novel and intuitive human interfaces for research and education in structural molecular biology utilizing 3D printing and augmented reality technologies.
Olson’s visualizations and animations have reached a broad audience through public venues such as the Disney EPCOT center, PBS television, and a number of art and science museum exhibits around the world.