May 5 2016
Researchers from the Scientific Research Institute of Physical-Chemical Medicine, MIPT, the company M&S Decisions and the research department of Yandex have built a computer model of the interaction between different bacteria, and between bacteria and the gut wall. This has led them to explain how antibiotic-resistant microbes develop and spread; details of the study have been published in the journal PLOS One.
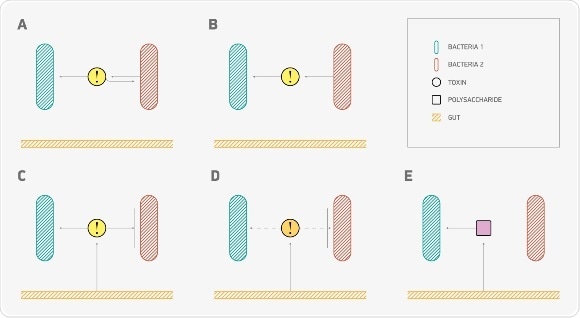
Feedbacks’ mechanisms. (A) Toxin-antitoxin system: bacterium produces toxins, which are harmful to itself and digestible to the other bacterial type. (B) Bacteria of one type control abundance of the other bacterial type. (A) and (B) situations have been also examined in inversed edge directions. (C) Gut produces toxins, which are harmful to one bacterial type and digestible to the other one (toxin’s type is the same as in A case). (D) Gut controls abundance of bacterial species by producing toxins against one bacterial type (1 or 2). (E) Gut produces digestible substrates for bacteria of type 2. Source: MIPT press office
The human intestine contains trillions of different bacteria, which together are called the microbiome. Bacteria protect us from harmful microorganisms, produce digestive enzymes, and help the immune system to function normally. Many diseases, such as obesity, Crohn’s disease, colon cancer, and other inflammatory processes are associated with a change in the gut microbiome. The researchers built a model of the interaction between two types of bacteria and the intestine and they determined what happens when antibiotics are taken that kill a large number of microorganisms.
“In biomedical science, we are now actively gathering information that is extremely important for bioinformatics – information which is enabling us to build models of complex systems. In our study, we used a simple modelling method – Agent Based Modelling (ABM) – to recreate the processes involving bacteria that take place in the gut and explain some interesting effects when resistivity occurs,” said Dmitry Alexeev, Deputy Head of MIPT’s Laboratory of Systems Biology and the corresponding author of the study.
The researchers were interested in a number of important issues – firstly, the speed at which the number of bacteria was restored after antibiotic therapy. Secondly, the scientists were interested in finding what proportion of bacteria was not affected by antibiotics, and thirdly the model described in detail the process of feedback between bacteria and the intestinal wall. The intestinal wall actively absorbs certain substances and produces others, which affects the number of bacteria and their “state of health”: if the numbers of bacteria in these processes are altered, it is difficult to obtain proper results.
The substances produced by bacteria and the intestinal walls were one of the main focal points in the new model. These compounds, which are formed by the bacterial fermentation of carbohydrates, may perform different functions at once: e.g. they can be toxins for one type of bacteria, and “food” for another type. If certain bacteria begin to produce too much of a substance that is poisonous to them, their numbers automatically decrease – scientists call this negative feedback. The intestinal wall can also produce either compounds that are harmful to bacteria (but not to the intestines themselves!), or carbohydrates that microorganisms can digest as nutrients.
The model also enabled the scientists to observe the location of strains in the gut and how their position changes after various substances are produced by the intestinal wall or bacteria. The visualization of the model showed that the spatial structure is a key factor that helps bacteria to survive and adapt to the changing conditions of their environment.
Using the model, the researchers were able to prove that even after antibiotic therapy there may be more bacteria that are sensitive to the effects of the antibiotic drug than those that are resistant to it. This conclusion may seem contradictory, but it is in fact true: the resistance to antibiotics comes at a price to the microorganisms. Antibiotics work by disrupting the biochemical mechanisms that are of vital importance to the function of the microbes (e.g. blocking protein synthesis or respiratory enzymes), which means they can only survive with major changes to the structure of key proteins, or by producing molecules that neutralize the drug. By altering their biochemical properties or using up resources on additional chemical reactions, bacteria inevitably lose out on efficiency: they consume “food” slower, divide slower and therefore it is certainly not the case that these microbes will be able to drive out ordinary microorganisms. The fact that we now have increasing numbers of antibiotic-resistant strains is important both from a scientific, as well as a medical point of view.
Observing the position of bacteria in the gut on the model also enabled the researchers to explain the possible advantage of resistant strains, which could lose out to less resistant, but faster rivals. The researchers say that regions dominated by one particular type of bacteria play a key role here – in these regions the “slow but resistant” strains simply suppress the competition. Feedback mechanisms and the production and absorption of substances in the gut wall also have an effect on the ratio of resistant and sensitive bacteria. The more feedback mechanisms involved, the slower the reduction in the number of sensitive bacteria, while resistant strains die faster. The researchers believe that further studies in this area will be useful to understand the fundamental causes of the emergence of antibiotic-resistant bacteria, and to develop drugs targeted at pathogens.
Biophysics and the system approach
The intestinal microbiome is not traditionally an area studied at MIPT: since its founding, MIPT has been known as a centre for training physicists with a focus on applied and engineering subjects such as microelectronics, the aerospace industry, and information technology. However, in recent years the institute has begun to look more closely at the field of life science – and not only in the faculties of biological and medical physics, but also in the “purely physical” Department of General and Applied Physics which now has its very own biophysics section. A new biopharmaceutical department has been built on campus, new laboratories have been opened to study the structure of proteins, and specialized conferences have also been held at the department.
It is no coincidence that physicists are moving towards biology. Modern biology and medicine involve both complex physical and chemical methods (e.g. detailed simulations of protein molecules are obtained using X-ray lasers and accelerators), and a vast amount of data, which can only be analysed using powerful computer equipment and intelligently optimised algorithms. The combination of high volumes of data with biology has even led to the development of a separate discipline – bioinformatics.