Jul 14 2016
DNA polymerases are the “Xerox machines” that replicate our DNA. They must work with great precision to keep errors from creeping into our genes. In spite of this precision, they still accept building blocks that have been coupled to large proteins, as a group of German scientists reports in the journal Angewandte Chemie. Based on this fact, the team has developed detection systems for genotyping DNA and RNA that can be evaluated by the naked eye. This method may allow for new diagnostic tools for use in the field.
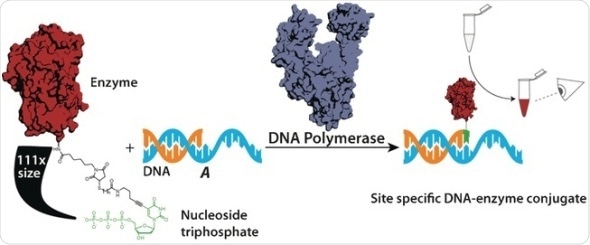
DNA polymerases read the information from a single strand of DNA and build up the complementary strand, one nucleotide at a time. Although they need to “recognize” the shape of the nucleotides to be included, the also accept slightly modified nucleotides. This is used in biotechnology for adding markers. A team led by Andreas Marx at the University of Konstanz have now demonstrated that highly selective DNA polymerases also incorporate nucleotides that have been attached to large proteins like horseradish peroxidase — even though this “cargo” is over 100 times larger than the nucleotide itself. The only requirement is that a linking group between the protein and the nucleotide must keep a minimum distance between them.
This astonishing result was used by the researchers to develop detection systems that make the presence of a specific DNA or RNA detectable with the naked eye. This may make it possible to detect gene fragments that are tied to genetic diseases or cancer.
To make this kind of test, a short DNA segment (the primer), complementary to the sequence being detected, is attached to a support. If a sample being tested contains the DNA segment in question, the corresponding single strand (known as the template) binds to the primer. A single incorrect base prevents this binding. Subsequently, a DNA polymerase and a chimera made from a nucleotide and horseradish peroxidase are added. If a template is bound to the primer, the polymerase is activated and couples the carefully selected nucleotide chimera to the template. Any unbound chimera is washed away and a colorless reagent is added. This reagent is converted to a brownish dye by the horseradish peroxidase. The intensity of the color can be used to estimate the concentration of the DNA fragment being detected with the naked eye. It was possible to detect as little as 1 fmol.
It was also possible to do a very targeted search for defined point mutations. For this procedure, the primer must end just before the relevant site. The template then binds to the primer, but the nucleotide–enzyme chimera only couples on if the template has the target base at this location.
By using a different polymerase, the researchers were also able to detect specific bacterial RNA, a process that is very useful for the identification of pathogens.
Because the polymerases used in these processes are active at room temperature, sophisticated laboratory equipment like thermocyclers is not necessary. It is therefore possible to carry out such tests by the patient’s side or in remote locations.