Sponsored Content by PittconJan 23 2017
An interview with Professor Jonathan Sweedler, University of Illinois at Urbana-Champaign, conducted by April Cashin-Garbutt, MA (Cantab)
Please can you give a brief introduction to your research and cell-to-cell signaling in the brain?
My group develops approaches to study cell-to-cell signaling in the brain – how the cells of the brain talk to each other. The brain is heterogeneous, probably more so than any other organ in our body, and many of its functions depend on the unique characteristics of these cells.
Memory, for example, by definition, arises partly because of the connections between individual cells. And so, in order to understand questions about memory, or how the brain controls behavior, or even age-old questions about consciousness, the brain must be studied at the cellular level. However, measuring the chemistry at the level of individual cells is difficult.
A microliter of brain tissue has over 100,000 cells, and this microliter will contain many different types of cells, such as astrocytes, oligodendrocytes and neurons. If one measures the proteins or metabolites in this microliter, you would learn about a hypothetical average cell. You would not gain information on the contents of neurons or other unique cell types.
What we're really after, and what many neuroscientists want to know, concerns cell heterogeneity and chemistry. We hope to determine the chemicals that neurons in the brain use to talk to each other, and how these change during learning and memory, and during different behaviors.
We could then address how these change with addiction, mental diseases, etc. Understanding the single cell level will help us answer these global questions.
Mass Spectrometry for Brain Research
Why do many cell-to-cell signaling molecules in the brain remain poorly characterized?
Most people know about serotonin because of its role in depression and the use of serotonin reuptake inhibitors, and also they know about dopamine in terms of attention deficit disorder or Parkinson's, but there are lots of other transmitters and neuromodulators too.
Scientists have found neurotransmitters, like glutamate, serotonin and dopamine, that are used by a significant fraction of cells in the brain; however, if there are transmitters that are used by only rare cells, we may not know about them.
Consider, for example, neuropeptides and neurohormones, which are very important for changing our mood or, from a cellular perspective, changing the firing threshold of specific neuronal circuits. Sometimes a neuropeptide may only be found in a rare cell, located in a specific part of the brain where it is only present in 1 in a 1,000 cells; this makes it easy to overlook.
Another characteristic that can make neurotransmitters difficult to characterize is that some of these molecules are formed by the actions of multiple enzymes. This means that you can't look at a particular form of a neuropeptide and point to a specific gene that makes it, because there are a lot of enzymes that help in the process.
Because of this, techniques like transcriptomics allow you to evaluate the potential neuropeptides involved in a cell, but measurements must be made to confirm whether the specific biologically active peptide form is present.
For a lot of these reasons, we're still discovering brain cell-to-cell signaling molecules. Additionally, researchers are still trying to come up with the entire parts list of the brain, so not just cell-to-cell signaling molecules, but all of the lipids and fatty acids and metabolites in the brain. These are dynamic and are not completely characterized.
What bioanalytical techniques are now available to investigate individual neurons and small brain regions?
To make a measurement of the contents of a test tube and investigate what's in it, mass spectrometry and liquid chromatography mass spectrometry are commonly employed. We also must consider whether a technique can be downscaled to work at the single cell level.
Separation techniques, such as capillary scale liquid chromatography and capillary electrophoresis, work well at this level. Capillary electrophoresis is well suited for taking individual cells and separating what's in them, and mass spectrometry can be used to characterize the individual components.
Accordingly, we have developed a range of capillary electrophoresis mass spectrometry approaches to sample the cytoplasm in individual cells and characterize the major metabolites within a cell.
Direct mass spectrometry is another approach, and this is perhaps the most common single cell techniques we use. In one embodiment, we scatter 10,000 cells onto a microscope slide; then we make the cells fluorescent and image the slide via fluorescence microscopy to determine their x-y locations on the slide.
We feed those locations into our mass spectrometer, and so sample only from the cells and probe what's in them.
This type of high-throughput, single cell profiling allows us to look at 10,000 cells at a time to find some of the major molecules within them. This is one of the highlights of the talk I'll be giving at Pittcon: how we can take a sample from a brain region and use this approach to probe the neuropeptides and some of the lipids and small molecules in those cells.
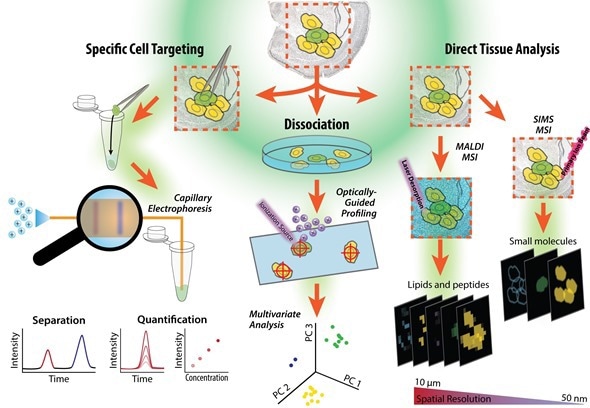
Figure illustrating workflow of a tissue sample, from cell isolation to our single cell measurements based on tissue imaging, single cell profiling and capillary electrophoresis-mass spectrometry. The cells can be dispersed onto a microscope slide and probed, or specific cells can be selected and assayed using capillary electrophoresis mass spectrometry. Figure prepared by Troy Comi from the Sweedler laboratory at the University of Illinois at Urbana-Champaign.
One thing that's really exciting about this technique is that it leaves about 80% of the material behind. After the measurement, if we find that a few of the cells are particularly interesting, or have something unusual, for example, if cell number 940 has a different profile that we didn't expect, we know its exact x-y location on the slide. We can then follow up and do immunofluorescence on that specific cell, or we can even isolate the cell and investigate it with capillary electrophoresis-mass spectrometry.
This high-throughput profiling can then be followed up with other techniques in order to learn more information about the rare cells we come across.
Which approaches will you be focusing on in your upcoming talk at Pittcon 2017?
At Pittcon I will be talking about high-throughput single cell profiling using two flavors of mass spectroscopy:
- Matrix-assisted laser desorption ionization (MALDI) mass spectrometry
- Secondary Ion mass spectrometry (SIMS)
Both of these techniques can be used for profiling large cellular populations. I will also be talking about rare cells and using capillary electrophoresis mass spectrometry and capillary electrophoresis laser induced fluorescence to investigate the contents of these cells. Why so many flavors of mass spectrometry? One of the reasons is that each one gives us a little bit different information on the chemicals in the cells.
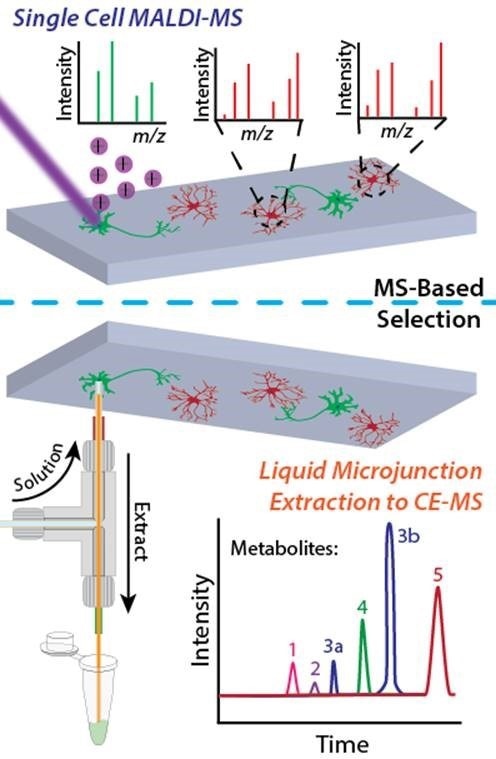
The Sweedler group's single cell metabolomics workflow combines direct mass spectrometry profiling and capillary electrophoresis-based mass spectrometry in a two-tiered process. Image prepared by Troy Comi and Elizabeth Neumann from the Sweedler laboratory at the University of Illinois at Urbana-Champaign.
What is the key to successful measurement?
The important point is that our measurements work and are robust. The most common reasons for failure involve experimental design and sampling issues. It’s important to remember, I am talking about the sampling protocols and the design of complex animal experiments.
We work with animals from across the animal kingdom, from comb jellies with their simple brains, to sea slugs, and mammals such as rodents, in the effort to link neurochemistry, physiology and behavior.
The key to successful measurements depends on how you design the experiment. If you're trying to design an experiment to assay what changes occur during drug escalation for a drug abuse study, or during disease or even during memory formation, you have to make sure that the experiment is carried out in a way that allows the dynamic chemistry in the cells to be measured.
Another thing to remember is that some of the molecules of interest, such as neuropeptides and small molecules, degrade very quickly after death, and so choosing the correct experimental protocol to retrieve the single cells is extremely important. We've had to come up with ways to preserve the cells to ensure compatibility with our measurement approaches.
An example of a cell preservation technique, which a lot of neuroscientists would suggest, is cell fixation. However, this is a way of turning the cell into plastic, as it crosslinks many of the cell’s molecules.
While it preserves the cell, it degrades the resulting mass spectrometry data. We spend a lot of time validating and designing our protocols, because if a sample is degraded, no amount of analytical measurement can rescue it.
I'm being a little general because sometimes the experiments are done locally, and sometimes they're performed at distant collaborator laboratories; factors like this will change how we go about things.
How much do you vary your approach for the type of sample being assayed and metabolites being measured?
It varies between cases, but to give an example, when looking for rare cells, sometimes we use a sample where the cells of interest are fluorescent model. We then try to find these fluorescent cells, either in a tissue or dispersed on a slide, or even by using fluorescent cell sorting, and then we analyze them via mass spectrometry.
Moving away from the brain, we have isolated cells from pancreatic islets and investigate the peptides in the different islet cell types: alpha, beta, gamma, or delta cells.
Our sampling approaches depend on whether it's a rare cell and the type of molecule we want to measure; for example, if we want to know the lipids in a cell, we make very different measurements than we do for neuropeptides.
Transcriptomics, the most common form of single cell measurement, provides a fairly complete set of the transcripts in the cell. When we do mass spectrometry on cells, we use the terms metabolomics or peptidomics, but the results are far from complete.
It's not really "omics"-scale. For example, if there are 20,000 metabolites in the brain, and we do metabolomics in a brain region, we can measure hundreds to a few thousand metabolites.
These represent only a few percent of the metabolites. The specific metabolites (or proteins or peptides) that are characterized depend on the specific mass spec platform and operating protocols used.
If we know in advance that we're looking for a particular subset of cellular metabolites, such as the hydrophobic versus the hydrophilic, we adjust our protocols accordingly. We have to optimize our methods more frequently because unfortunately, the measurements we make are not global or omics-scale as they are with DNA and RNA.
How important are single cell assays to your work?
My group is evenly split between technology development and neuroscience measurements. For the technology development, about 70% is single cell measurement, and for the biological measurements it is about 30% single cell measurements.
This is an exciting time for brain research. All around the world, in Europe, the Americas, and in Asia, there are huge initiatives to understand the brain. In America, it's the BRAIN Initiative, which is run through the National Institutes of Health (NIH), the National Science Foundation (NSF), and other organization.
One of the goals is to understand cell heterogeneity and build a cell census of the brain. Within the US, there are several BRAIN initiatives related to creating single cell measurements, and we're are involved with these efforts.
What technology advances are in the pipeline and which companies are working on these?
We use mass spectrometry for most of our measurements. For our imaging and cell profiling work, higher acquisition speed and better detectability are important. We need new instruments that provide more information from a smaller sample.
The obvious players are: Bruker, Agilent, Sciex, Thermo Scientific, Waters, among others. They're coming up with better instruments that give you more information from smaller samples.
If you're performing single cell profiling with MALDI, for example, Bruker is creating faster mass spectrometers that provide more information from a greater number of cells. We are also trying to integrate single cell mass spectrometry with other approaches such as vibrational spectroscopy single cell transcriptomics.
The other area that is rapidly evolving involves the way we handle cells. There is an ever increasing range of approaches for cell sampling, dispersing cells and hyphenating aspects of cell sampling with microfluidics.
A lot of these approaches are being created by academic research groups, but I also see small companies that create and offer these technologies. The combination of new mass spectrometers, better informatics and improved cell sampling suggests that the age of single cell chemical characterization has a bright future.
What impact do you think these new technologies will have on your ability to uncover the complex chemical mosaic of the brain?
Let me highlight two areas. First, new technologies and new protocols are allowing us to look for a greater range of chemistry in the brain. These advances have been rapid and enabling. For an example of an area outside of my research, I have been working with several groups in developing vibrational spectroscopy.
They can now image a brain slice with infrared and Raman spectroscopy at cellular resolution, getting complete vibrational spectra on thousands of cells. Now we can do infrared or Raman imaging on a brain slice, and then perform mass spectrometry afterwards on that same slice, which provides unprecedented chemical detail.
Another exciting advance is that these technologies are becoming more robust, so that more groups are able to use them. As an example, my group has helped teach a course on single cell measurements at the Cold Spring Harbor Laboratory.
When we teach people how to do single cell sampling and mass spectrometry, a lot of them will go back to their own local mass spec facility; in the past, the facility managers would tell potential users that single cell measurements would not work on their facility instruments. Now more of these facilities are willing to try (and succeed) with these types of measurements.
What do you think the future holds for pinpointing key cellular players in physiological and pathological processes in the brain?
Two things. One, there are billions of cells and we're only measuring thousands. We need to become more global in our measurements. Second, we often find unusual peaks/masses in a cell but confirming what these unexpected compounds are remains a challenge.
Additionally, a lot of the single cell measurements have been qualitative. We need to know what's in the cell, and what's in the cell that changes based on disease and/or a physiological process. We're getting to the point where our goal is to increase the quantitative details. Adding more robust quantitative information to these types of measurements will really increase their impact.
Where can readers find more information?
Further reading/references:
- Single Cell Peptide Heterogeneity of Rat Islets of Langerhans. Jansson ET, Comi TJ, Rubakhin SS, Sweedler JV. ACS Chem Biol. 2016,11(9):2588-95. 10.1021/acschembio.6b00602.
- Classification of Large Cellular Populations and Discovery of Rare Cells Using Single Cell Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Ong TH, Kissick DJ, Jansson ET, Comi TJ, Romanova EV, Rubakhin SS, Sweedler JV. Anal Chem. 2015, 87(14):7036-42. 10.1021/acs.analchem.5b01557.
- Small-volume Analysis of Cell-Cell Signaling Molecules in the Brain. Romanova EV, Aerts JT, Croushore CA, Sweedler JV. Neuropsychopharmacology. 2014, 39(1):50-64. 10.1038/npp.2013.145. Review.
- Single-cell Analysis at the Threshold, Chen X, Love JC, Navin NE, Pachter L, Stubbington MJ, Svensson V, Sweedler JV, Teichmann SA. Nat Biotechnol. 2016, 34(11):1111-1118. 10.1038/nbt.3721. Feature.
About Professor Jonathan Sweedler
 Jonathan Sweedler received his Ph.D. in Chemistry from the University of Arizona in 1988 and spent several years at Stanford before moving to the University of Illinois at Urbana-Champaign in 1991, where he has been ever since.
Jonathan Sweedler received his Ph.D. in Chemistry from the University of Arizona in 1988 and spent several years at Stanford before moving to the University of Illinois at Urbana-Champaign in 1991, where he has been ever since.
At Illinois, he is currently the James R. Eiszner Family Endowed Chair in Chemistry, Director of the School of Chemical Sciences, and affiliated with the Institute of Genomic Biology and the Beckman Institute for Advanced Science and Technology.
His research interests focus on developing new approaches for assaying small volume samples, and in applying these methods to study novel interactions between cells. These analytical approaches include capillary separations, micro- and nanofluidics, miniaturized separations, mass spectrometry and nuclear magnetic resonance.
He has used these tools to characterize small molecules and peptides in a range of animal models across metazoan life, and in samples as small as individual cells and cellular domains. Recent work includes the development of a series of high throughput mass spectrometry approaches for characterizing tens of thousands of individual cells.
Sweedler, with large international teams of biologists and technologists, has performed comprehensive interrogation of the genome, transcriptome and peptidome in a range of animal models to uncover the signaling peptides and pathways involved in a wide range of functions and behaviors.
Sweedler has published more than 370 manuscripts and presented 480 invited lectures. He has received numerous awards including the American Chemical Society (ACS) Analytical Division Arthur Findeis Award, the Benedetti-Pichler Award in Microanalysis, the Gill Prize in Neuroscience, the Instrumentation Award from the Analytical Division of the ACS, the Pittsburgh Analytical Chemistry Award, the ACS Award in Analytical Chemistry and the ANACHEM Award.
He is a fellow of both the American Association for the Advancement of Science and the American Chemical Society. He is currently the Editor-in-Chief for Analytical Chemistry.
About Pittcon
 Pittcon® is a registered trademark of The Pittsburgh Conference on Analytical Chemistry and Applied Spectroscopy, a Pennsylvania non-profit organization. Co-sponsored by the Spectroscopy Society of Pittsburgh and the Society for Analytical Chemists of Pittsburgh, Pittcon is the premier annual conference and exposition on laboratory science.
Pittcon® is a registered trademark of The Pittsburgh Conference on Analytical Chemistry and Applied Spectroscopy, a Pennsylvania non-profit organization. Co-sponsored by the Spectroscopy Society of Pittsburgh and the Society for Analytical Chemists of Pittsburgh, Pittcon is the premier annual conference and exposition on laboratory science.
Proceeds from Pittcon fund science education and outreach at all levels, kindergarten through adult. Pittcon donates more than a million dollars a year to provide financial and administrative support for various science outreach activities including science equipment grants, research grants, scholarships and internships for students, awards to teachers and professors, and grants to public science centers, libraries and museums.
Visit pittcon.org for more information.