Chromatrap® has published a new article that describes a detailed genome wide comparison of native and crosslinked chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) using the company's patented solid state spin column technology.
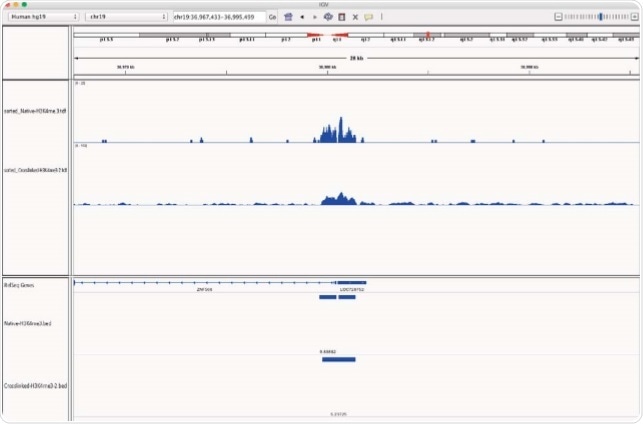
Chromatrap® supply high quality N-ChIP-seq and X-ChIP-seq kits. This article compares the enrichment of the histone H3K4me3 modification signal across the whole genome, from chromatin prepared by both methods (native vs cross link). The histone mark H3K4me3 was chosen for this study to highlight the sensitivity and efficiency of the Chromatrap® spin column technology for both native and cross-linked ChIP-seq as it is associated with ‘open’ transcription start sites and gene activation and is present in euchromatin throughout the genome.
Data is presented which demonstrates strong and comparable enrichment of H3K4me3 from both Native and Cross-linked chromatin. The study highlights that both Native and Cross-linked chromatin preparation methods are compatible with Chromatrap® extraction and capture technology. Providing excellent data both these methods ensure superior H3K4me3 enrichment followed by high throughput sequencing. In addition, the study shows high quality FASTQC metrics and significant genome wide peak enrichment levels are obtainable using both assay technologies.
To download a copy of the application note ‘Chromatrap® ChIP-seq from native and cross-linked chromatin‘ please visit https://www.chromatrap.com/activity/.